Team:UCSF/Project/Arsenal
From 2010.igem.org
Jasonpark1 (Talk | contribs) m (→BETTER ARSENAL) |
(→BETTER ARSENAL) |
||
| (17 intermediate revisions not shown) | |||
| Line 18: | Line 18: | ||
| - | ===''' | + | ==='''Better Arsenal'''=== |
| - | + | <br> | |
| - | <br | + | |
[[Image:UCSFnewgranuleagents-06.jpg|center|500px|Immune synapse]] | [[Image:UCSFnewgranuleagents-06.jpg|center|500px|Immune synapse]] | ||
| - | <br | + | <br> |
'''Goal:''' | '''Goal:''' | ||
| Line 32: | Line 31: | ||
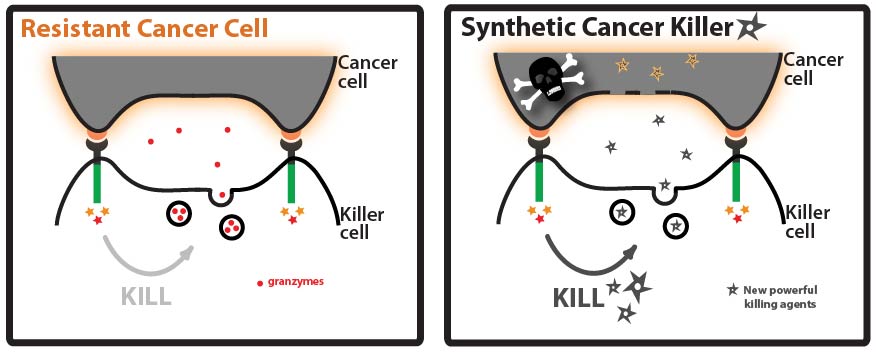
One way that cancer cells can evade the immune system is by evolving resistance to the arsenal of cytotoxic proteins found in killer cells’ granules. Our goal for this summer was to direct a model protein, green fluorescent protein (GFP), to killer cells’ granules by identifying and fusing granule localization “address” tags to GFP. This would be an important first step toward loading granules with novel cytotoxic proteins against which cancer cells would likely be defenseless! | One way that cancer cells can evade the immune system is by evolving resistance to the arsenal of cytotoxic proteins found in killer cells’ granules. Our goal for this summer was to direct a model protein, green fluorescent protein (GFP), to killer cells’ granules by identifying and fusing granule localization “address” tags to GFP. This would be an important first step toward loading granules with novel cytotoxic proteins against which cancer cells would likely be defenseless! | ||
While it is not completely understood how killer cells send proteins to cytotoxic granules, we were able to learn a great deal about the “address” tags of various proteins that are sent to granules by searching and reading the literature. We designed a number of parts using these tags and made devices by fusing them in various combinations to GFP. | While it is not completely understood how killer cells send proteins to cytotoxic granules, we were able to learn a great deal about the “address” tags of various proteins that are sent to granules by searching and reading the literature. We designed a number of parts using these tags and made devices by fusing them in various combinations to GFP. | ||
| + | |||
| + | <br><br> | ||
===Brief Introduction=== | ===Brief Introduction=== | ||
| Line 50: | Line 51: | ||
Granzymes are transported through the MPR-dependent pathway<html><a href="#References">[5]</a></html>.The signal peptide of the granzymes get cleaved upon entering the Endo reticulum. However, they are inactive in the Endo reticulum in the Endo reticulum and the golgi. Only after they reached granule, the N-terminal dipeptide can be cleaved off by DPPI aka cathepsin C, this leaves behind the active protein which begins with the conserved IIGG sequence. For Example, the N-terminal of Granzyme B is QPILLLLAFLLLPRADAGEIIGG underlined Amino acids are the signal sequence. The GE in italic is the dipeptide which inhibits the activity of granzyme B. In the granule, GE will get cleaved off by cathepsin C, and then granzyme B is activated. | Granzymes are transported through the MPR-dependent pathway<html><a href="#References">[5]</a></html>.The signal peptide of the granzymes get cleaved upon entering the Endo reticulum. However, they are inactive in the Endo reticulum in the Endo reticulum and the golgi. Only after they reached granule, the N-terminal dipeptide can be cleaved off by DPPI aka cathepsin C, this leaves behind the active protein which begins with the conserved IIGG sequence. For Example, the N-terminal of Granzyme B is QPILLLLAFLLLPRADAGEIIGG underlined Amino acids are the signal sequence. The GE in italic is the dipeptide which inhibits the activity of granzyme B. In the granule, GE will get cleaved off by cathepsin C, and then granzyme B is activated. | ||
| + | |||
| + | <br><br> | ||
===Directing GFP to killer cells' granules by fusing "address" tags=== | ===Directing GFP to killer cells' granules by fusing "address" tags=== | ||
| Line 109: | Line 112: | ||
GILT stands for “glycosylation-independent lysosomal targeting”, it actually uses a truncation of insulin-like grow factor II (IGF II). The Cation Independent mannose phosphate receptor has a IGF II binding motif. Since CI-MPR can also go to the cell membrane, its natural function is to internalize the circulating IGF II proteins and send them to the lysosome for degradation, thus regulating IGF II concentration. Therapy of lysosome storage disease takes advantage of the IGF II, and fuse exogenous lysosomal enzymes to the GILT tag. These genetically modified enzymes can be taken up by patient cells to fulfill some deficient function. We use the GILT tag to deliver endogenous proteins to lysosome, which also looks feasible. | GILT stands for “glycosylation-independent lysosomal targeting”, it actually uses a truncation of insulin-like grow factor II (IGF II). The Cation Independent mannose phosphate receptor has a IGF II binding motif. Since CI-MPR can also go to the cell membrane, its natural function is to internalize the circulating IGF II proteins and send them to the lysosome for degradation, thus regulating IGF II concentration. Therapy of lysosome storage disease takes advantage of the IGF II, and fuse exogenous lysosomal enzymes to the GILT tag. These genetically modified enzymes can be taken up by patient cells to fulfill some deficient function. We use the GILT tag to deliver endogenous proteins to lysosome, which also looks feasible. | ||
| + | |||
| + | <br><br> | ||
===Cloning Strategy=== | ===Cloning Strategy=== | ||
| Line 125: | Line 130: | ||
After we get all the AB, BC and CD parts, we were able the assemble them in different combination and make devices for testing. | After we get all the AB, BC and CD parts, we were able the assemble them in different combination and make devices for testing. | ||
| + | |||
| + | <br><br> | ||
===Device List=== | ===Device List=== | ||
{|cellspacing="0" | {|cellspacing="0" | ||
| - | | | + | |'''#'''||'''[N-terminal Granzyme sequence]-[cargo]-[C-terminal “address” tag]'''||'''Plasmid Name''' |
|- | |- | ||
|1||GRZM B-EGFP-CDMPR||pCPL_056 | |1||GRZM B-EGFP-CDMPR||pCPL_056 | ||
| Line 141: | Line 148: | ||
|5||GRZM B-EGFP-Y TAIL||pCPL_060 | |5||GRZM B-EGFP-Y TAIL||pCPL_060 | ||
|- | |- | ||
| - | |6||GRZM B-EGFP-STOP||pCPL_061 | + | |6||GRZM B-EGFP-STOP||pCPL_061 |
|- | |- | ||
|7||GRZM M-EGFP-CDMPR||pCPL_062 | |7||GRZM M-EGFP-CDMPR||pCPL_062 | ||
| Line 161: | Line 168: | ||
===Testing the Constructs=== | ===Testing the Constructs=== | ||
| - | In this part of our project we used Amaxa electroporation to transfect NKL cells (an NK cell line) with plasmids containing our devices. Then we grew cells | + | In this part of our project we used Amaxa electroporation to transfect NKL cells (an NK cell line) with plasmids containing our devices. Then we grew cells and started to select for cells expressing our construct for our assay by selecting with the selection agent G418. The cells used for the assay were mixed populations as the selection was not complete. |
Our assay consisted of using fluorescence microscopy to determine how well we were able to target EGFP to the granules. Before microscopy, we first stained our NKL cell membranes with the membrane Vybrant DiI stain and then with the Lysotracker Blue lysosomal stain (killer cell granules are specialized lysosomes). After our initial assay we saw that the EGFP wasn’t fluorescing well, which we attributed to quenching due to the low pH levels of the granules, or degradation due to other lysosomal proteins. To address this issue we decided to fixed and permeabilized our cells, and stained the EGFP inside with a fluorescent anti-GFP antibody. This showed much clearer results. | Our assay consisted of using fluorescence microscopy to determine how well we were able to target EGFP to the granules. Before microscopy, we first stained our NKL cell membranes with the membrane Vybrant DiI stain and then with the Lysotracker Blue lysosomal stain (killer cell granules are specialized lysosomes). After our initial assay we saw that the EGFP wasn’t fluorescing well, which we attributed to quenching due to the low pH levels of the granules, or degradation due to other lysosomal proteins. To address this issue we decided to fixed and permeabilized our cells, and stained the EGFP inside with a fluorescent anti-GFP antibody. This showed much clearer results. | ||
| - | + | <br><br> | |
===Results=== | ===Results=== | ||
| Line 194: | Line 201: | ||
We were able to observe evidence of granular anti-GFP staining and workable granule staining in samples pCPL_056, pCPL_059, pCPL_060, and pCPL_061. (See summary table below.) In some samples, the granular anti-GFP staining and granule staining appear to be well-colocalized, suggesting that EGFP was loaded into the killer cell granules. | We were able to observe evidence of granular anti-GFP staining and workable granule staining in samples pCPL_056, pCPL_059, pCPL_060, and pCPL_061. (See summary table below.) In some samples, the granular anti-GFP staining and granule staining appear to be well-colocalized, suggesting that EGFP was loaded into the killer cell granules. | ||
| - | SUMMARY OF RESULTS | + | |
| + | '''SUMMARY OF RESULTS''' | ||
(See below for more details) | (See below for more details) | ||
colocalization stats images ready for wiki | colocalization stats images ready for wiki | ||
| - | {| | + | {|cellspacing="0" |
! Construct | ! Construct | ||
! GFP looks granular | ! GFP looks granular | ||
| Line 261: | Line 269: | ||
|N/A | |N/A | ||
|} | |} | ||
| + | |||
Here are the visual results for each sample: | Here are the visual results for each sample: | ||
| Line 268: | Line 277: | ||
This is our best result. The anti-GFP stain (green) is well colocalized with the Granule stain (red). It also appears to be well centralized in the cell which seems to show that it is stably located in the correct granules, since it is not being sent out of the cell. | This is our best result. The anti-GFP stain (green) is well colocalized with the Granule stain (red). It also appears to be well centralized in the cell which seems to show that it is stably located in the correct granules, since it is not being sent out of the cell. | ||
| - | + | <br><br> | |
[[Image:UCSF_59002.jpg]] | [[Image:UCSF_59002.jpg]] | ||
In this image we can see granular looking anti-GFP staining (green). However, it is located near the cell membrane and does not seem to overlap with the granule stain (red). | In this image we can see granular looking anti-GFP staining (green). However, it is located near the cell membrane and does not seem to overlap with the granule stain (red). | ||
| - | + | <br><br> | |
[[Image:UCSF_61001.jpg]] | [[Image:UCSF_61001.jpg]] | ||
| - | This image also shows well colocalized anti-GFP (green) and granules (red) (the yellow part represnts the heavily overlapped area) the results here are less clear than in image 56003, but nevertheless show centralized, colocalized anti-GFP and Granule stain | + | This image also shows well colocalized anti-GFP (green) and granules (red) (the yellow part represnts the heavily overlapped area) the results here are less clear than in image 56003, but nevertheless show centralized, colocalized anti-GFP and Granule stain. |
| - | + | <br><br> | |
[[Image:UCSF_61002.jpg]] | [[Image:UCSF_61002.jpg]] | ||
As you can see in this image, the granular anti-GFP is mostly at the edge of the membrane. The granule stain seems to slightly overlap the anti-GFP in some places, but is largely not colocalized with it. Since this construct only has the granzyme signal sequence we hypothesized that it would serve as a control for localization. | As you can see in this image, the granular anti-GFP is mostly at the edge of the membrane. The granule stain seems to slightly overlap the anti-GFP in some places, but is largely not colocalized with it. Since this construct only has the granzyme signal sequence we hypothesized that it would serve as a control for localization. | ||
| - | + | <br><br> | |
[[Image:UCSF_pMax_GFP.jpg]] | [[Image:UCSF_pMax_GFP.jpg]] | ||
In this image we have stained the pMAX-GFP with anti-GFP. As you can see, since the GFP is expressed throughout the cell, the stain is also everywhere. Unlike the EGFP constructs, this image has no granular GFP, but rather an entirely lite up cell. | In this image we have stained the pMAX-GFP with anti-GFP. As you can see, since the GFP is expressed throughout the cell, the stain is also everywhere. Unlike the EGFP constructs, this image has no granular GFP, but rather an entirely lite up cell. | ||
| + | <br><br><br> | ||
===Future application=== | ===Future application=== | ||
| - | + | <br /> | |
| + | [[Image:UCSFnewgranuleagents-07.jpg|center|600px]] | ||
| + | <br /> | ||
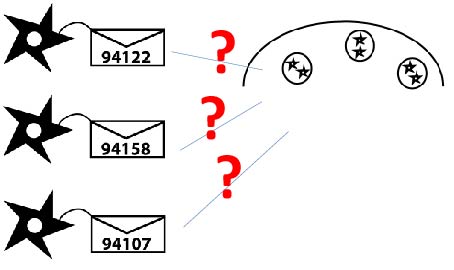
We are encouraged by our preliminary results suggesting that we can successfully target the model protein EGFP into killer cell granules. With a few more tests we can fully optimize the process and figure out what combination of N- and C- terminal address tags work the best. Then, we can use the best combinations of address tags to route novel cytotoxic cargo into the granules for cancer killing. Some of the various ideas we have explored for cargo that we could load into granules include: proteins important for normal regulation such as p53, and the cytochrome and BL family proteins; Stronger killing agents, such as TNF and Par-4. | We are encouraged by our preliminary results suggesting that we can successfully target the model protein EGFP into killer cell granules. With a few more tests we can fully optimize the process and figure out what combination of N- and C- terminal address tags work the best. Then, we can use the best combinations of address tags to route novel cytotoxic cargo into the granules for cancer killing. Some of the various ideas we have explored for cargo that we could load into granules include: proteins important for normal regulation such as p53, and the cytochrome and BL family proteins; Stronger killing agents, such as TNF and Par-4. | ||
We can also use it as a new means for delivering existing cancer therapies, such as growth inhibitors, with more specificity. Once this process has been further investigated and fine tuned, it will offer a new revolutionary means of combating cancer and creating more powerful killer cells. | We can also use it as a new means for delivering existing cancer therapies, such as growth inhibitors, with more specificity. Once this process has been further investigated and fine tuned, it will offer a new revolutionary means of combating cancer and creating more powerful killer cells. | ||
| - | + | <br> | |
| + | <br> | ||
===References=== | ===References=== | ||
| Line 372: | Line 385: | ||
doi:10.1016/j.bbamcr.2008.10.016 | doi:10.1016/j.bbamcr.2008.10.016 | ||
| - | www.cimr.cam.ac.uk/investigators/robinson/docs/Bonifacino%2009.pdf | + | http://www.cimr.cam.ac.uk/investigators/robinson/docs/Bonifacino%2009.pdf |
Latest revision as of 03:37, 28 October 2010
|
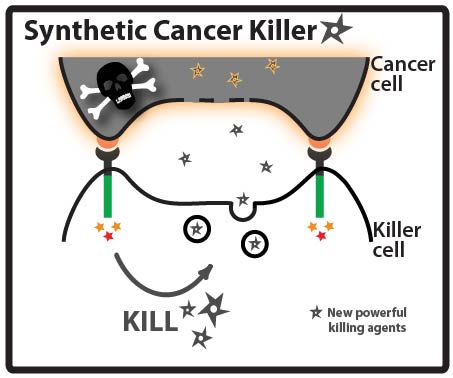
Better Arsenal
Goal: Direct a model protein, green fluorescent protein (GFP), to killer cells’ granules by identifying and fusing granule localization “address” tags to the protein. Approach: One way that cancer cells can evade the immune system is by evolving resistance to the arsenal of cytotoxic proteins found in killer cells’ granules. Our goal for this summer was to direct a model protein, green fluorescent protein (GFP), to killer cells’ granules by identifying and fusing granule localization “address” tags to GFP. This would be an important first step toward loading granules with novel cytotoxic proteins against which cancer cells would likely be defenseless! While it is not completely understood how killer cells send proteins to cytotoxic granules, we were able to learn a great deal about the “address” tags of various proteins that are sent to granules by searching and reading the literature. We designed a number of parts using these tags and made devices by fusing them in various combinations to GFP.
Brief Introduction
The address tags of the proteins utilize the cellular transportation system to go to different compartments of the cell. It is like mails, although there are addresses on them, they themselves will not go to the address; it depends on the post system to pick them up and deliver them to the right place. So what’s the transportation system of the cell like? They are made of transmembrane proteins. The rumen sides of the transporter proteins can recognize and bind to the address tags on cargo proteins. The cytoplasmic side of the transporter proteins contains specific motifs. When the golgi bubbles out, different bubbles contains different sets of transporter proteins whose cytoplasmic motifs will decide where the bubble should go (The mechanism will not be discussed here). Now let’s take a look at how granule resident proteins are sorted to the granule. By transporter, the sorting pathways can be divided into Mannose Phosphate Receptor (MPR)-dependent pathway and MPR-independent pathways. MPR-dependent pathways use MPR as the transporter. And the address tag is mannose-6-phosphate. Proteins transported in this pathway are glycosylated with mannose-6-phosphate groups, these groups serve as the address tag. The rumen side of the MPR can bind to the mannose-6-phosphate motif (We can see the from the naming of MPR). The cytoplasmic side of MPR contains “DXXLL” motif, which decides that MPR will be sorted to lysosome/granule. MPR-independent pathways, from the name, use transporters other than MPR. It may or may not depend on the mannose-6-phosphate tag on the cargo. This includes sortilin, LIMPII, LRP;[2][3][4]the addressing tags involved are not very clear. Granzymes are transported through the MPR-dependent pathway[5].The signal peptide of the granzymes get cleaved upon entering the Endo reticulum. However, they are inactive in the Endo reticulum in the Endo reticulum and the golgi. Only after they reached granule, the N-terminal dipeptide can be cleaved off by DPPI aka cathepsin C, this leaves behind the active protein which begins with the conserved IIGG sequence. For Example, the N-terminal of Granzyme B is QPILLLLAFLLLPRADAGEIIGG underlined Amino acids are the signal sequence. The GE in italic is the dipeptide which inhibits the activity of granzyme B. In the granule, GE will get cleaved off by cathepsin C, and then granzyme B is activated.
Directing GFP to killer cells' granules by fusing "address" tags
We hypothesized that fusing address tags from proteins that are naturally sent to the granule may work to send GFP to the granule as well.
Proteins should be translocated into the endoplasmic reticulum (ER) first in order to be further sorted to granule. Since it is not sure whether the signal sequence would play some role in the granule sorting, we decided to use the signal sequences from the granzymes. Each Granzyme has an individual signal peptide sequence that delivers it to the Endo Reticulum. Every signal sequence is distinct in size and amino acid content.
We have two strategies to send our cargo from the endoplasmic reticulum (ER) to the granule: 1. Directly fuse our cargo to the granule specific transporters. 2. Fuse our cargo to some address tag which can bind to the transporter.
The proteins listed below all have granule localization motif on their cytoplasmic tails.
Y-motif: In a previous work[6], scientists delivered TNFR1 to the granule by fusing it to the CD63 trans-membrane and cytoplasmic fragment. The mechanism is that the CD63 protein is a lysosome resident protein, it has signal sequence “SIRSGYEVM” on its cytoplasmic end, which contains the YXXØ motif (X represents any Amino Acid, Ø stands for hydrophobic amino acid). Similarly, other protein cargos can also be fused to the CD63 protein to be transported to the granule. MPR: Many of the normal granule resident proteins in the cell are glycosylated and form a mannose-6-phosphate in the Golgi. MPR(mannose phosphate receptor) are membrane receptors for the mannose-6-phosphate motif. The cytoplasmic motif “DXXLL” of the MPR decided that it will be sorted to the granule together with the cargos bound. We make it another candidate to fuse our cargo to. LIMP-II & LAMP1: Lysosomal membrane proteins have been proposed to reach lysosomes by two pathways referred to as “direct” and “indirect”. In the direct pathway, lysosomal membrane proteins are transported intracellularly from the TGN to either early or late endosomes and then to lysosomes without ever appearing at the cell surface. In contrast, the indirect pathway involves constitutive transport from the TGN to the plasma membrane, followed by internalization into early endosomes and eventual delivery to late endosomes and lysosomes. Members of the LAMP/LIMP class are the preeminent example of proteins thought to traffic via the direct pathway, that’s why we picked LIMP-II and LAMP1. We hope that by employing the direct pathway we can reduce the leakiness of our cargo to extracellular compartment. Sortilin: Sortilin is identified to be an alternative receptor than MPR in transporting lysosomal proteins. Sortilin has been reported to mediate lysosomal trafficking of prosaposin and acid sphingomyelinase. The cytoplasmic tail “GYHDDSDEDLL” contains “DXXLL” motif.
Strategy 2
However, by looking at the works on the treatment of lysosomal storage diseases, we found a glycosylation independent motif that binds to MPR[7].
GILT stands for “glycosylation-independent lysosomal targeting”, it actually uses a truncation of insulin-like grow factor II (IGF II). The Cation Independent mannose phosphate receptor has a IGF II binding motif. Since CI-MPR can also go to the cell membrane, its natural function is to internalize the circulating IGF II proteins and send them to the lysosome for degradation, thus regulating IGF II concentration. Therapy of lysosome storage disease takes advantage of the IGF II, and fuse exogenous lysosomal enzymes to the GILT tag. These genetically modified enzymes can be taken up by patient cells to fulfill some deficient function. We use the GILT tag to deliver endogenous proteins to lysosome, which also looks feasible.
Cloning StrategyWe used the BBF RFC 28 strategy to make fusion protein. Signal sequences are made into AB parts the cargo (GFP) is made into BC part and all the granule localization adress tags are made into CD parts. After assembling the AB-BC and CD part, we can get the constructs we want and express them in CTL cells. Since the signal sequences and the granule localization sequences are all short in length, we decided to make the basic AB and BC parts fused together through overlap extension PCR-based methods used in gene synthesis. These methods are robust for assembling oligos of 40-50nt into sequences up to and greater than 3kb. (Protocols from personal communication, Jason Park). We used the web-based software Helix System DNAworks (http://helixweb.nih.gov/dnaworks/) for human codon-optimization and oligonucleotide parsing. After we get all the AB, BC and CD parts, we were able the assemble them in different combination and make devices for testing.
Device List
Testing the ConstructsIn this part of our project we used Amaxa electroporation to transfect NKL cells (an NK cell line) with plasmids containing our devices. Then we grew cells and started to select for cells expressing our construct for our assay by selecting with the selection agent G418. The cells used for the assay were mixed populations as the selection was not complete. Our assay consisted of using fluorescence microscopy to determine how well we were able to target EGFP to the granules. Before microscopy, we first stained our NKL cell membranes with the membrane Vybrant DiI stain and then with the Lysotracker Blue lysosomal stain (killer cell granules are specialized lysosomes). After our initial assay we saw that the EGFP wasn’t fluorescing well, which we attributed to quenching due to the low pH levels of the granules, or degradation due to other lysosomal proteins. To address this issue we decided to fixed and permeabilized our cells, and stained the EGFP inside with a fluorescent anti-GFP antibody. This showed much clearer results.
ResultsWe were able to test six of our devices in the assay: pCPL_056, pCPL_059, pCPL_060, pCPL_061, pCPL_063, and pCPL_067. We also had two negative controls for comparison:
We analyzed this condition because we wanted to have a negative control for auto-fluorescence in the cell line. However, we had some problems because the background staining intensity of the fluorescent anti-GFP antibody was different from the experimental samples. We hypothesized that this was because of the different health of these cells, which had not been growing under G418 selection.
This control is the same cell line (NKL) transfected with the pMAX-GFP plasmid. pMAX-GFP is a powerful expression vector for GFP available from Lonza. Because it only expresses GFP, and doesn’t contain the N-terminal granzyme signals or the C-terminal sorting tags, the GFP will not be sent to the granules. The purpose of this control is to illustrate the difference between GFP left in the cytosol and GFP localized into the granules.
- At times, staining with the anti-GFP antibody did not appear to be uniform between all of the different samples. This was likely a technical error and results probably would be improved with more practice. - Staining the granules with the lysosomal stain Lysotracker Blue (Invitrogen Molecular Probes) was often problematic. Most cells appeared to have high levels of non-specific background staining in the cytoplasm, especially after the cells were fixed and permeabilized. In addition, we were told that fluorescent dyes in the blue part of the light spectrum are often more challenging to visualize (more noise). However, we were able to observe reasonable Lysotracker Blue staining in some of our images. In some of our images, we had to independently adjust the LUTs settings for the Lysotracker to visualize the staining in some of our images (noted in the results tables below). We performed the image processing function “Smoothing” (NIS-Elements AR software) on all of our images (identical settings for all images). We were able to observe evidence of granular anti-GFP staining and workable granule staining in samples pCPL_056, pCPL_059, pCPL_060, and pCPL_061. (See summary table below.) In some samples, the granular anti-GFP staining and granule staining appear to be well-colocalized, suggesting that EGFP was loaded into the killer cell granules.
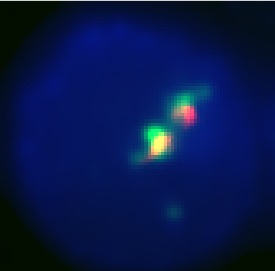
This is our best result. The anti-GFP stain (green) is well colocalized with the Granule stain (red). It also appears to be well centralized in the cell which seems to show that it is stably located in the correct granules, since it is not being sent out of the cell.
In this image we can see granular looking anti-GFP staining (green). However, it is located near the cell membrane and does not seem to overlap with the granule stain (red).
This image also shows well colocalized anti-GFP (green) and granules (red) (the yellow part represnts the heavily overlapped area) the results here are less clear than in image 56003, but nevertheless show centralized, colocalized anti-GFP and Granule stain.
As you can see in this image, the granular anti-GFP is mostly at the edge of the membrane. The granule stain seems to slightly overlap the anti-GFP in some places, but is largely not colocalized with it. Since this construct only has the granzyme signal sequence we hypothesized that it would serve as a control for localization.
In this image we have stained the pMAX-GFP with anti-GFP. As you can see, since the GFP is expressed throughout the cell, the stain is also everywhere. Unlike the EGFP constructs, this image has no granular GFP, but rather an entirely lite up cell.
Future application
References1. L is for lytic granules: lysosomes that kill Page LJ, Darmon AJ, Uellner R, Griffiths GM. Biochim Biophys Acta. 1998 Feb 4;1401(2):146-56. http://www.ncbi.nlm.nih.gov/pubmed/9531970
Lefrancois S, Zeng J, Hassan AJ, Canuel M, Morales CR. EMBO J. 2003 Dec 15;22(24):6430-7. http://www.ncbi.nlm.nih.gov/pubmed/14657016
Reczek D, Schwake M, Schr?der J, Hughes H, Blanz J, Jin X, Brondyk W, Van Patten S, Edmunds T, Saftig P. Cell. 2007 Nov 16;131(4):770-83. http://www.cell.com/abstract/S0092-8674%2807%2901290-1
Hiesberger T, Hüttler S, Rohlmann A, Schneider W, Sandhoff K, Herz J. EMBO J. 1998 Aug 17;17(16):4617-25. http://www.ncbi.nlm.nih.gov/pmc/articles/PMC1170791/
J Cell Biol. 1993 Feb;120(4):885-96. http://www.ncbi.nlm.nih.gov/pubmed/8432729
Hansson M, Gao Y, Rosén H, Tapper H, Olsson I. Eur Cytokine Netw. 2004 Jul-Sep;15(3):167-76. http://www.ncbi.nlm.nih.gov/pubmed/15542440
LeBowitz JH, Grubb JH, Maga JA, Schmiel DH, Vogler C, Sly WS. Proc Natl Acad Sci U S A. 2004 Mar 2;101(9):3083-8. Epub 2004 Feb 19. http://www.ncbi.nlm.nih.gov/pubmed/14976248
References for sorting signals:
Sorting of lysosomal proteins Thomas Braulke and Juan S. Bonifacino doi:10.1016/j.bbamcr.2008.10.016 http://www.cimr.cam.ac.uk/investigators/robinson/docs/Bonifacino%2009.pdf
|
 "
"