Team:UCL London/Project Modeling
From 2010.igem.org
| Line 35: | Line 35: | ||
==CIRCUIT DESIGN== | ==CIRCUIT DESIGN== | ||
| + | '''Many thanks to Dr. Yuhong Zhou for all her help in developing the mathematical equations''' | ||
| + | The design of a biological system should include all the information needed to describe what is happening within the cell (Carlson, 2010). As in with our case, the numerical simulation of all the biochemical reactions identified, was to describe the system, and specific values such as the rate constant, are unknown, leading to the simulation exploring a wide variety of possible values from literature. Even if we did have particular values for the variables appearing in the model, it would still be impossible to make accurate predictions about its behaviour in real life. According to Carslon (2010), the line between model and simulation becomes blurred when the only way to assess a particular model is depending upon the simulation of the model. However, again the design stage with the simulation differs significantly from the actual model. This is where the difficulty lays, as scientists and engineers, work on projects together by repeatedly comparing simulation and measurement through experiments. The Bristol team for this year iGEM competition, has assisted us, in the production of a standard simulation, with the potential to run multiple simulations using their BioSim programme, to explore the behaviour of the model, over a range of conditions. However, the experiments, in terms of the laboratory experiments of the physical system, to explore the model behaviour, must be carried out and hopefully with the aid of theoretical behavioural descriptions from textbooks, we can produce quantitative predictions. | ||
| + | The SyntheticBiology.org website defines Synthetic Biology as: | ||
| + | |||
| + | A) the design and construction of new biological parts, devices, and systems, and | ||
| + | |||
| + | B) the re-design of existing, natural biological systems for useful purposes. | ||
| + | |||
| + | The cartoon description used to describe the protein expression as a result of the transcription of DNA into RNA by polymerase which is in turn translated into protein. The ribosomes, which are not included in the animation, play an important role in the production of the proteins, according to the “central dogma”, defined by Francis Crick in 1957 that information flows from DNA to RNA to protein. | ||
| + | |||
| + | The challenges around the biological systems are the following, and summarise the scope of the formation of the iGEM competition: | ||
| + | |||
| + | 1. Defining the device physics of molecular components | ||
| + | 2. Building and simulating models | ||
| + | 3. Using defined components from a standard library (biobricks) | ||
| + | 4. Building new biological entities based on quantitatively predictive designs | ||
| + | |||
| + | The genetic switch, that we aimed to build in the bacterium E.coli, was to be independent of the application of a chemical signal, such as IPTG, that would induce the cells to start producing the protein. The bacterium, when the switch is on, is predicted to make red fluorescent protein; when the switch is off, the protein is no longer made and any remaining protein decays. | ||
| + | |||
| + | The fundamental that the genetic circuit works on, is that a signal from outside the cell, that is when the oxygen spike appears, declaring the low oxygen concentration present, induces the cell to begin producing the proteins. Traditionally, this was done with the manual addition by the operator of IPTG to the cells. The employment of inducible promoters, i.e. pNark, that works by interfering with a repressor protein, LacI, a protein that inhibits transcription by binding to the promoter. The polymerase, T7RNA P, is located downstream of the pNark, where it binds to DNA and begins transcription. The repressor protein, LacI, blocks the access to the pLacI promoter, because it is prevented by the pNark promoter. Therefore the role of the pNark inducible promoter is double: to accept the signal for cell induction and production of mRNA and proteins through the loop and also to inhibit the repressor binding to DNA. The addition of IPTG into the system, will cause the induction of the pLacI promoter which was repressed by the LacI proteins during the duration of the positive feedback loop and will be the regulator. The anti-PA binds to the promoter activator and reversibly stops the positive loop and hence, transcription terminates. | ||
| + | |||
| + | According to Purnick and Weiss (2009), the first wave of synthetic biology includes the combination of promoters, RBS (ribosome binding sites) and transcriptional repressors for the formation of functional modules e.g. controlled protein expression. Such a novel circuit design was based on principles like the creation and analysis of a genetic circuit, the experimental evaluation of the circuit and the combatibility of the results with literature. Moreover, they refer to directed evolution, which is about the optimisation of biobricks and the system i.e. the biological circuit. | ||
Revision as of 08:50, 21 October 2010
PROJECT HYPOXON - MODELLING
“The process defines the product” is the motto we always have in mind when designing the route for the production of a compound.
Biopharmaceuticals are commonly synthesized using E. coli as production chassis. Typically, the production of biopharmaceuticals is triggered by the addition of an induction agent, often IPTG, during the bioprocess. Imagine if the bacteria as we know it today could drive protein expression by linking the hypoxia condition to oxygen sensitive promoters within the cells. Synthetic biology through biochemical engineering can create the potential at which Escherichia coli cells- and we all know how harmful to humans can specific strains be- used in a twisted yet beautifully way as an expression system for the production of biopharmaceuticals. The project is based on the engineering modification of the cells and the construction of a self-induced biological circuit that allows an automated protein expression.
We all remember the Swine flu pandemic that outbreak that occurred about a year ago. The production of such a great amount of pharmaceuticals in such a short time to cover the needs of the infected people was only based on a well planned engineering scale-up equations and an established protocol. Therefore, in order to create a product and with regards to our project, a pharmaceutical drug, there is the need to establish the conditions that will enable the effective development of the process and the efficient and most economically viable product. By decreasing the cost of the project, it only means that it is delivered to the patient at a lower cost. By producing naturally occurring drugs at a large scale, avoiding as much as possible chemicals that can cause damage to the cells or side-effects of the final product it only means that a safer product has been manufactured and it is ready for consumption by the patient.
Biological engineering is yet indeed in the early years of its life- if it would be an innovative product it would be on the introduction phase, where there is around 2% of innovators and followers of that particular idea. What about the rest though? How do we make it possible for others to understand and moreover how do we make the whole idea approachable to others? This project had to be translated into the actual benefits that the society could gain from this. Therefore, as more “mature” engineering fields exist like mechanical and electrical, the project was “expressed” in those terms. For example the standard computational programmes SolidWorks and Verilog are powerful tools for the expression of equipment and circuits respectively in the engineering fields mentioned above. Comparatively engineering toolboxes that will enable the visual construction of models, using those programmes for the needs of a synthetic biology, will enable a quantitative understanding of how parts of a biological system when come together behave in different manners. Through this combination it is believed that the whole perspective of Synthetic Biology will be transformed (Carlson, 2009). The construction of a database with well characterised genetic parts, with assigned mathematically abstracted functions that can be modelled in a software and turning electronic circuits into molecule representations via DNA synthesis could lead to a more economically and social value of what Synthetic Biology projects can offer. The currently available computational tools for the representation of genetic networks, include Antimony, Athena, BioJade, GenoCAD, OptCircuit, SynBioSS and many more software tools which were constructed to integrate all the relevant aspects of a biological system (Purnick and Weiss, 2009).
According to Carlson (2010), the revenues from genetic modifications of biological systems in USA back in 2007 was around 2% of the GDP (gross domestic product). These products included drugs, crops, materials, industrial enzymes and fuels. The Parliament Office of Science and Technology (2008) posted that the global market for DNA sequencing technology and services, in 2006, as estimated by the US Department of Energy was over $7 billion. This rapid revenue growth from the sales of biotechnological products, suggests the evolution of a new market. The necessity to improve current biological technologies and aid the advancement of establishing them is still an issue. Large scale projects have demonstrated social and economic implications, such as the GM crops and the political, social and ethical implications it raised as a result of the introduction of Genetic Modifications as a new technological approach (Gregory and Jay Lock, 2008). The challenges, when it comes to the complexity of the biological systems and the application of them in our life are still influenced by the public policy, safety and biosecurity which suggest that although Biology is technology, and is in fact the oldest technology, Synthetic Biology is only at the beginning. Based on the uncertainty that some features in biology carry, the optimisation of such algorithms is still not reliable. For example, E.coli is a quite simple biological organism and is used as a chassis for a wide range of biological expressions; however, it still contains many uncharacterized genes (Carlson, 2010).
One of the goals of this project is to be able to take it from bench top to commercialisation of the technology and to optimise the conditions at a pilot scale prior to moving to large quantities production, at low cost and at higher yield. This could mean access to medication for people who could not have otherwise afforded it. In fact, many affected families live with less than $1 a day leading to the main question: How can these people get treatment since they can only spend a few pennies a day on drugs and the per capita health budget is only a few US dollars per year? (Carlson, 2010) According to WHO (World Health Organisation) the disease harms whole society by decreasing the GDP of the country. In particular (Gallup and Sachs, 2001) malaria affected countries reduces GDP growth by 1.3% per year while WHO reported that malaria-free countries have a threefold higher GDP per capita compared to those countries. From synthetic biology to biochemical engineering and from the genetic modifications of an E.Coli cell to the growth of that cell in order to achieve protein expression we aim to build a synthetic project where several parts come together and create a protocol for the production of pharmaceuticals. That’s not the whole story though. Our aim goes further into the modification of bacteria and accordingly other systems such as yeast or mammalian cell culture, the establishment of the optimum conditions to achieve their growth and expression of the desired product, downstream processing that should be adjusted to fit the needs of each system with the final goal to scale-up the process to cover the needs of large populations. What if, through an organised and well pictured project, where all the parts, like LEGOS, fitted together in all the possible ways through the synthetic biology game and one day we could come across to that specific gene modification that will enable the fight of cancer or hepatitis or even HIV? What if? Just imagine that.
We tried to predict the models that would enable such visualization with the hope that one day there will be specific genetic engineering software with its own database of biobricks and tools used for the random or algorithmic construction of biological parts that when assembled in reality can create a better biological system than already existed.
“It is only through learning how the parts work, and how they work together, that we will truly be able to produce engineered biological systems” Robert H. Carlson
The aim of adopting modelling tools is to enable us to make right predictions with regards to our project and to develop a standard protocol to be used as the basis for translating the construction of a biological circuit such as the bacterial (E.coli) circuit, to other expression systems for the production of novel biopharmaceuticals for the treatment of major diseases.
The modelling section is divided into 5 main sections for simplicity: 1. Circuit Construction
2. Bioprocess Flowsheet Development
3. Fermentation set-up: Mechanical Design of the Fermenter & Materials
4. Large-scale considerations: Economic Evaluation, Health & Safety Analysis, Waste Management & HAZOP studies
CIRCUIT DESIGN
Many thanks to Dr. Yuhong Zhou for all her help in developing the mathematical equations
The design of a biological system should include all the information needed to describe what is happening within the cell (Carlson, 2010). As in with our case, the numerical simulation of all the biochemical reactions identified, was to describe the system, and specific values such as the rate constant, are unknown, leading to the simulation exploring a wide variety of possible values from literature. Even if we did have particular values for the variables appearing in the model, it would still be impossible to make accurate predictions about its behaviour in real life. According to Carslon (2010), the line between model and simulation becomes blurred when the only way to assess a particular model is depending upon the simulation of the model. However, again the design stage with the simulation differs significantly from the actual model. This is where the difficulty lays, as scientists and engineers, work on projects together by repeatedly comparing simulation and measurement through experiments. The Bristol team for this year iGEM competition, has assisted us, in the production of a standard simulation, with the potential to run multiple simulations using their BioSim programme, to explore the behaviour of the model, over a range of conditions. However, the experiments, in terms of the laboratory experiments of the physical system, to explore the model behaviour, must be carried out and hopefully with the aid of theoretical behavioural descriptions from textbooks, we can produce quantitative predictions.
The SyntheticBiology.org website defines Synthetic Biology as:
A) the design and construction of new biological parts, devices, and systems, and
B) the re-design of existing, natural biological systems for useful purposes.
The cartoon description used to describe the protein expression as a result of the transcription of DNA into RNA by polymerase which is in turn translated into protein. The ribosomes, which are not included in the animation, play an important role in the production of the proteins, according to the “central dogma”, defined by Francis Crick in 1957 that information flows from DNA to RNA to protein.
The challenges around the biological systems are the following, and summarise the scope of the formation of the iGEM competition:
1. Defining the device physics of molecular components 2. Building and simulating models 3. Using defined components from a standard library (biobricks) 4. Building new biological entities based on quantitatively predictive designs
The genetic switch, that we aimed to build in the bacterium E.coli, was to be independent of the application of a chemical signal, such as IPTG, that would induce the cells to start producing the protein. The bacterium, when the switch is on, is predicted to make red fluorescent protein; when the switch is off, the protein is no longer made and any remaining protein decays.
The fundamental that the genetic circuit works on, is that a signal from outside the cell, that is when the oxygen spike appears, declaring the low oxygen concentration present, induces the cell to begin producing the proteins. Traditionally, this was done with the manual addition by the operator of IPTG to the cells. The employment of inducible promoters, i.e. pNark, that works by interfering with a repressor protein, LacI, a protein that inhibits transcription by binding to the promoter. The polymerase, T7RNA P, is located downstream of the pNark, where it binds to DNA and begins transcription. The repressor protein, LacI, blocks the access to the pLacI promoter, because it is prevented by the pNark promoter. Therefore the role of the pNark inducible promoter is double: to accept the signal for cell induction and production of mRNA and proteins through the loop and also to inhibit the repressor binding to DNA. The addition of IPTG into the system, will cause the induction of the pLacI promoter which was repressed by the LacI proteins during the duration of the positive feedback loop and will be the regulator. The anti-PA binds to the promoter activator and reversibly stops the positive loop and hence, transcription terminates.
According to Purnick and Weiss (2009), the first wave of synthetic biology includes the combination of promoters, RBS (ribosome binding sites) and transcriptional repressors for the formation of functional modules e.g. controlled protein expression. Such a novel circuit design was based on principles like the creation and analysis of a genetic circuit, the experimental evaluation of the circuit and the combatibility of the results with literature. Moreover, they refer to directed evolution, which is about the optimisation of biobricks and the system i.e. the biological circuit.
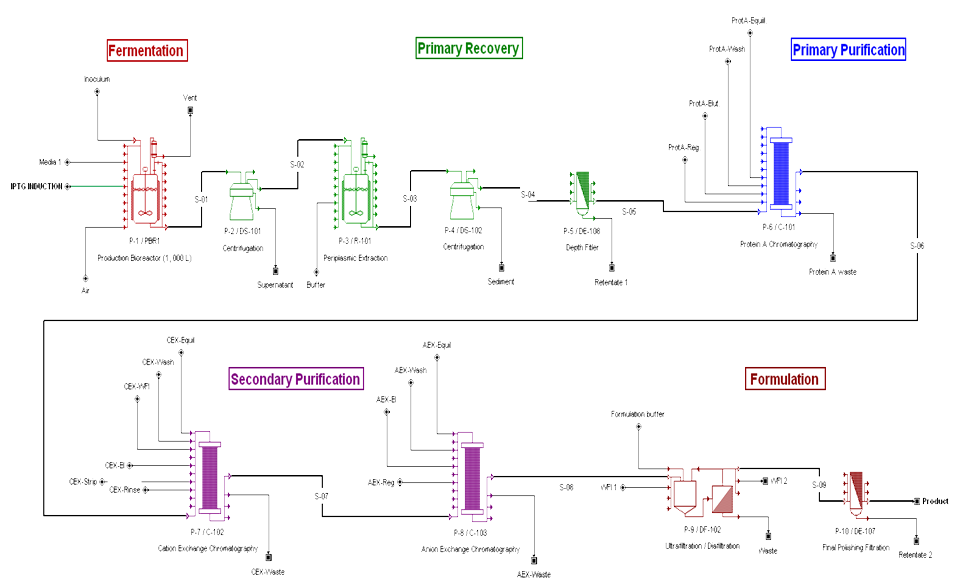
BIOPROCESS FLOWSHEET
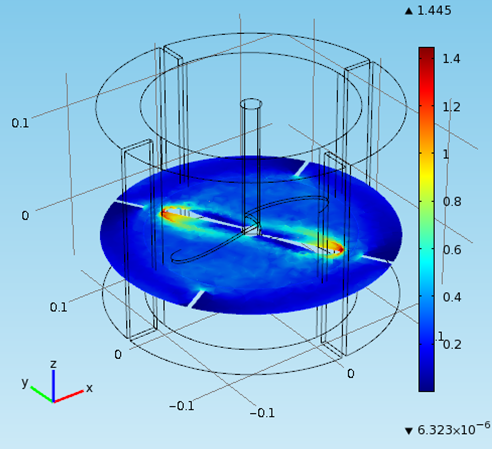
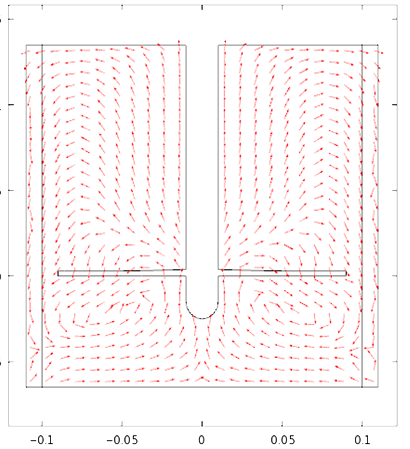
FERMENTER MECHANICAL DESIGN
LARGE SCALE CONSIDERATIONS
ECONOMICAL CONSIDERATIONS
HEALTH & SAFETY ANALYSIS
WASTE MANAGEMENT
HAZARD STUDIES
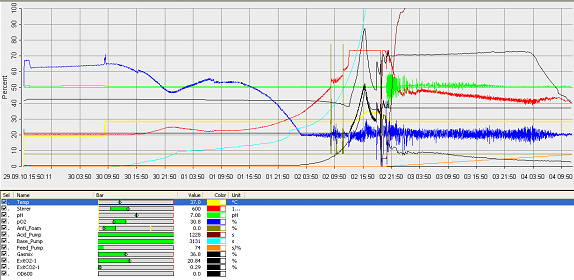
Fermentation


 "
"



 Twitter
Twitter Facebook
Facebook UCL
UCL Flickr
Flickr YouTube
YouTube