Team:Tokyo Tech/Project/Artificial Cooperation System/lux act rep
From 2010.igem.org
(→lux activation/repression part) |
|||
| (115 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
<div id="tf_menu"> | <div id="tf_menu"> | ||
| - | menu | + | <font size="5" color="#eb8300"><b><center>Project menu</center></b></font> |
| + | |||
| + | <center> | ||
| + | <table id="table-01"> | ||
| + | <tr> | ||
| + | <td>[[Team:Tokyo_Tech|1 Graphic abstract]]<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <td>2 Apple reporter<br> | ||
| + | :[[Team:Tokyo_Tech/Project/Apple_Reporter|2-1 Color]] | ||
| + | :[[Team:Tokyo_Tech/Project/Apple_Reporter2|2-2 Fragrance]] | ||
| + | </td> | ||
| + | <tr> | ||
| + | <th>[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System|3 Artificial Cooperation System]]<br> | ||
| + | :3-1 ''lux'' activation/repression promoter -YOU ARE HERE!- | ||
| + | :[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/Cm_assay|3-2 resistance gene activation device]] | ||
| + | :[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/luxI_assay|3-3 ''lux''I Assay]] | ||
| + | :[[Team:Tokyo_Tech/Project/Artificial_Cooperation_System/modeling|3-4 modeling]] | ||
| + | </th> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>[[Team:Tokyo_Tech/Project/wolf_coli|4 Wolf coli overview]]<br> | ||
| + | :[[Team:Tokyo_Tech/Project/wolf_coli/New_Series_of_PompC|4-1 New series of P''ompC'']] | ||
| + | :[[Team:Tokyo_Tech/Project/wolf_coli/lacIM1|4-2 lacIM1 for band-detect network ]] | ||
| + | :[[Team:Tokyo_Tech/Project/wolf_coli/System|4-3 Wolf coli system]] | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | </center> | ||
| + | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> <!-- tf_menu --> | </div> <!-- tf_menu --> | ||
| - | <div id="tf_SubWrapper"> | + | |
| - | < | + | <div id="tf_SubWrapper"> |
| - | = | + | <font size="5"><b>3-1 ''lux'' activation/repression promoter</b></font> |
| - | + | ==Abstract== | |
| + | In Artificial Cooperation System, two types of cells use quorum sensing to recognize population of the counterpart and to help the counterparts when they are dying. The quorum sensing in this system is regulated by AHL dependent transcriptional activation/repression. Therefore, we characterized activation/repression promoters. We examined the existing LuxR repression promoter which has never been characterized before in BioBrick registry. Even though the GFP expression was repressed in the presence of AHL, cell-growth rate decreased because of the overexpression of GFP occurred in the absence of AHL. For this reason, we designed and constructed a new repression promoter that regulates the transcription appropriately dependent on the signal input. | ||
| + | <br> | ||
| + | |||
| + | |||
| + | [[IMAGE:Tokyotech_plux_act_final.jpg|300px|left|thumb|fig.3-1-1 LuxR activation promoter assay (worked by Kitano Shohei & Eriko Uchikoshi)]] | ||
| + | |||
| + | [[IMAGE:Tokyotech_plux_rep_final.jpg|300px|left|thumb|fig.3-1-3 LuxR repression promoter assay (worked by Shohei Kitano & Eriko Uchikoshi)]] | ||
| + | |||
| + | ==Introduction== | ||
| + | |||
| + | In Synthetic Biology, transcription activation is used frequently. Transcription repression by using AHL is also important, however, the device which has delay for transcription/translation through inverter is used a lot in this case. We decided to experience LuxR repression promoter for the quick response of signal dependent repression.<br> | ||
| + | |||
| + | ==Result== | ||
| + | ===R0, characterization of R0062 (promoter activated by LuxR/3OC6HSL)=== | ||
| + | First, we characterized R0062, the well-known LuxR activation promoter in order to establish our Tokyo_Tech team experimental system for Artificial Cooperation System.<br> | ||
| + | The expression of GFP with 100nM 3OC6HSL around 30-folds increased comparing with the expression without 3OC6HSL.<br> | ||
| + | |||
| + | [[IMAGE:Tokyotech_plux_act_final.jpg|300px|left|thumb|fig.3-1-1 LuxR activation promoter assay (worked by Kitano Shohei & Eriko Uchikoshi)]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | We confirmed fluorescence intensity of LuxR activation promoter is dependent on 3OC6HSL concentration. The AHL concentration which shows half of maximam activity is less than 5nM. <br> | ||
| + | [[IMAGE:tokyotech_LuxR ractivatio promoter assay2.jpg|400px|left|thumb|fig.3-1-2 Fluorescence intensity dependent on the concentration of AHL (worked by Shohei Kitano & Eriko Uchikoshi)]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | ===R1, characterization of R0061 & K395008 (promoter repressed by LuxR/3OC6HSL)=== | ||
| + | |||
| + | [[IMAGE:Tokyotech_plux_rep_final.jpg|300px|left|thumb|fig.3-1-3 LuxR repression promoter assay (worked by Shohei Kitano & Eriko Uchikoshi)]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | ===R1-1, R0061 (promoter repressed by LuxR/3OC6HSL)=== | |
| + | Next, we characterized the existing part R0061, LuxR repression promoter. We examined whether the amount of transcription is appropriate when signal is off and how much this promoter represses. <br> | ||
| + | The expression of GFP with 100nM 3OC6HSL dropped to 1/3 comparing with the expression without 3OC6HSL. | ||
| - | === | + | ===R1-2, K395008 (promoter repressed by LuxR/3OC6HSL):R0061 weak=== |
| - | We confirmed | + | We confirmed R0061 and found increase of cells was inhibited due to a high level of expression although it is repressed by AHL. Therefore, we designed a new appropriate promoter by changing one base of R0061.<br> |
| - | + | The expression of GFP with 100nM 3OC6HSL dropped to 1/3 comparing with the expression without 3OC6HSL. We found the level of expression is appropriate and this promoter work as expected. | |
| - | == | + | ==Conclusion== |
| + | We designed the new promoter which is repressed LuxR/3OC6HSL complex by changing one base of existing promoter. We confirmed this promoter works as we expected.<br> | ||
| + | It is not so difficult to make the promoter which strength is between these two by designing. | ||
| - | === | + | ==Material & Methods== |
| - | + | ===M0, characterization of R0062 (promoter activated by LuxR/3OC6HSL)=== | |
| - | + | ===fluorescence intensity in the presence/absence of AHL === | |
| + | We constructed K395100 combining R0062 and K121013. K121013 is a promoter-less ''gfp'' reporter (rbs-''gfp''-ter-ter) on pSB6A1. S03119 is a LuxR generator which is regulated by PTetR, which is repressed by TetR. In this experiment, we don’t use TetR, so S03119 functions as a LuxR constitutive generator. The backbone of S03119 is pSB1A2, which is a high copy plasmid, so we changed the backbone from pSB1A2 to pSB3K3. | ||
| + | We used a fusion of PlacI<sup>q</sup> (I14032) to ''gfp'' (K121013) as a positive control and used promoterless ''gfp'' (K121013) as a negative control. | ||
| - | [[IMAGE: | + | [[IMAGE:Tokyotech_R0062assay_construction.png|400px]] |
| - | + | *samples | |
| - | + | #[Plux act - ''gfp''](BBa_K395100) on pSB6A1 + [PtetR – LuxR] on pSB3K3 | |
| + | #positive control: [PlacI<sup>q</sup>(constitutive promoter) - ''gfp''] on pSB6A1+ [PtetR – LuxR] on pSB3K3 | ||
| + | #negative control:. [promoterless - ''gfp''] on pSB6A1+ [PtetR – LuxR] on pSB3K3 | ||
| + | *Strain | ||
| + | DH5α | ||
| + | *protocol | ||
| + | #Prepare overnight culture. | ||
| + | #Take 30 ul of the overnight culture into LB + antibiotics (Amp + Kan). (→fresh culture) | ||
| + | #Incubate the fresh culture until the observed O.D. reaches around 0.60. | ||
| + | #Each sample was divided into 2. Prepare and add 3OC6HSL mixture to one, and add DMSO mixture to the other. The final concentration of 3OC6HSL is 100nM. | ||
| + | #Induction for 3 hours at 37°C. | ||
| + | #Fluorometer (FLA5200) and flow cytometry measurements for GFP expression. | ||
| - | === | + | ====fluorescence intensity dependent on [AHL]==== |
| - | + | *samples | |
| + | #[Plux act - ''gfp''](BBa_K395100) on pSB6A1 + [ptet – LuxR] on pSB3K3 | ||
| + | *Strain | ||
| + | DH5α | ||
| + | *protocol | ||
| + | #Prepare overnight culture. | ||
| + | #Take 30 ul of the overnight culture into LB + antibiotics (Amp + Kan). (→fresh culture) Prepare the same 7 tubes for each sample. | ||
| + | #Incubate the fresh culture until the observed O.D. reaches around 0.60. | ||
| + | #Each sample was divided into 2. Prepare and add 3OC6HSL mixture. The final concentration of 3OC6HSL is 1, 3, 5, 10, 30, 50, 100nM. | ||
| + | #Induction for 3 hours at 37°C. | ||
| + | #Fluorometer (FLA5200) and flow cytometry measurements for GFP expression. | ||
| - | + | ===M1, characterization of R0061 & K395008 (promoter repressed by LuxR/3OC6HSL)=== | |
| + | ====M1-1, characterization of R0061 (promoter repressed by LuxR/3OC6HSL)==== | ||
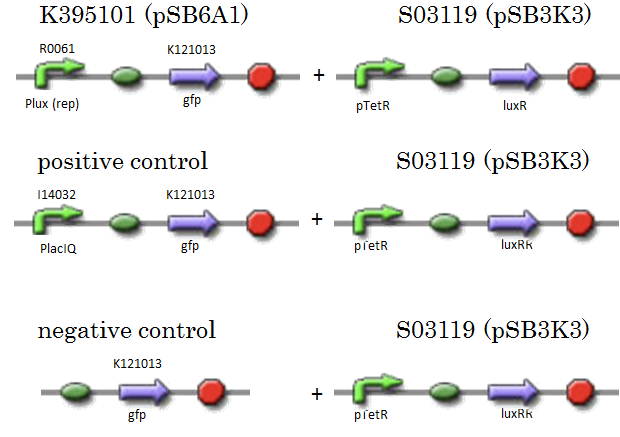
| - | + | We constructed K395101 combining R0061 and K121013, which is a promoter-less ''gfp'' reporter (rbs-''gfp''-ter-ter) on pSB6A1. S03119 is a LuxR generator which is repressed by TetR. In this experiment, we don’t use TetR, therefore, S03119 functions a LuxR constitutive generator. The backbone of S03119 is pSB1A2, which is a high copy plasmid, so we changed the backbone from pSB1A2 to pSB3K3. | |
| + | We used a fusion of PlacI<sup>q</sup> (I14032) to ''gfp'' (K121013) as a positive control and used promoterless ''gfp'' (K121013) as a negative control. | ||
| - | [[IMAGE: | + | [[IMAGE:Tokyotech_R0061assay_construction.png|400px]] |
| - | + | *'''samples''' | |
| - | + | #[Plux rep - ''gfp''](BBa_K395101) on pSB6A1 + [PtetR – LuxR] on pSB3K3 | |
| - | + | #positive control: [PlacI<sup>q</sup>(constitutive promoter) - ''gfp''] on pSB6A1+ [PtetR – LuxR] on pSB3K3 | |
| - | + | #negative control: [promoterless - ''gfp''] on pSB6A1+ [PtetR – LuxR] on pSB3K3 | |
| - | [ | + | *'''Strain''' |
| - | + | DH5α | |
| + | *'''protocol''' | ||
| + | #Prepare overnight culture. | ||
| + | #Take 30 ul of the overnight culture into LB + antibiotics (Amp + Kan).(→fresh culture) | ||
| + | #Incubate the fresh culture until the observed O.D. reaches around 0.80. | ||
| + | #Each sample was divided into 2. Prepare and add 3OC6HSL mixture to one, and add DMSO mixture to the other. The final concentration of AHL is 100nM. | ||
| + | #Induction for 2 hours at 37°C. | ||
| + | #Fluorometer (FLA5200) and flow cytometry measurements for GFP expression. | ||
| + | ====M1-2, characterization of K395008 (promoter repressed by LuxR/3OC6HSL):R0061 weak==== | ||
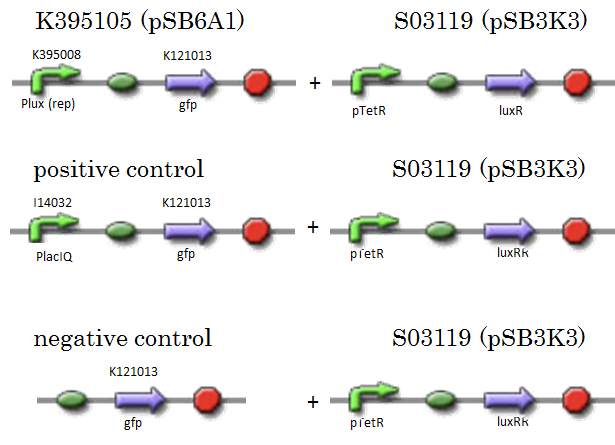
| + | We constructed K395105 combining K395008 and K121013. K121013 is a promoter-less ''gfp'' reporter (rbs-''gfp''-ter-ter) and this backbone is pSB6A1. Promoter of S03119 is PtetR, which is repressed by tetR. In this experiment, we don’t use TetR, so, S03119 functions a LuxR constitutive generator. The backbone of S03119 is pSB1A2, which is a high copy plasmid, so we changed the backbone from pSB1A2 to pSB3K3. | ||
| + | We used a fusion of PlacI<sup>q</sup> (I14032) to ''gfp'' (K121013) as a positive control and used promoterless ''gfp'' (K121013) as a negative control. | ||
| + | [[IMAGE:Tokyotech_K395008assay_construction.png|400px]] | ||
| + | *samples | ||
| + | #[R0061weak - ''gfp''](BBa_K395105) on pSB6A1 + [PtetR – LuxR] on pSB3K3 | ||
| + | #positive control: [PlacI<sup>q</sup>(constitutive promoter) - ''gfp''] on pSB6A1+ [PtetR – LuxR]) on pSB3K3 | ||
| + | #negative control: [promoterless - ''gfp''] on pSB6A1+ [PtetR – LuxR] on pSB3K3 | ||
| + | *Strain | ||
| + | DH5α | ||
| + | *protocol | ||
| + | #Prepare overnight culture. | ||
| + | #Take 30 ul of the overnight culture into LB + antibiotics (Amp + Kan). (→fresh culture) | ||
| + | #Incubate the fresh culture until the observed O.D. reaches around 0.60. | ||
| + | #Each sample was divided into 2. Prepare and add 3OC6HSL mixture to one, and add DMSO mixture to the other. The final concentration of 3OC6HSL is 100nM. | ||
| + | #Induction for 3 hours at 37°C. | ||
| + | #Fluorometer (FLA5200) and flow cytometry measurements for GFP expression. | ||
| + | ==Reference== | ||
| + | #KRISTI A. EGLAND & E. P. GREENBERG, Conversion of the Vibrio fischeri Transcriptional Activator LuxR, to a Repressor. JOURNAL OF BACTERIOLOGY, Feb. 2000, p. 805–811 | ||
Latest revision as of 03:58, 28 October 2010
3-1 lux activation/repression promoter
Contents |
Abstract
In Artificial Cooperation System, two types of cells use quorum sensing to recognize population of the counterpart and to help the counterparts when they are dying. The quorum sensing in this system is regulated by AHL dependent transcriptional activation/repression. Therefore, we characterized activation/repression promoters. We examined the existing LuxR repression promoter which has never been characterized before in BioBrick registry. Even though the GFP expression was repressed in the presence of AHL, cell-growth rate decreased because of the overexpression of GFP occurred in the absence of AHL. For this reason, we designed and constructed a new repression promoter that regulates the transcription appropriately dependent on the signal input.
Introduction
In Synthetic Biology, transcription activation is used frequently. Transcription repression by using AHL is also important, however, the device which has delay for transcription/translation through inverter is used a lot in this case. We decided to experience LuxR repression promoter for the quick response of signal dependent repression.
Result
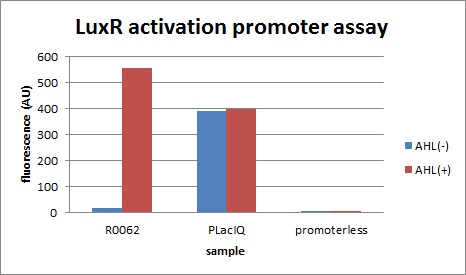
R0, characterization of R0062 (promoter activated by LuxR/3OC6HSL)
First, we characterized R0062, the well-known LuxR activation promoter in order to establish our Tokyo_Tech team experimental system for Artificial Cooperation System.
The expression of GFP with 100nM 3OC6HSL around 30-folds increased comparing with the expression without 3OC6HSL.
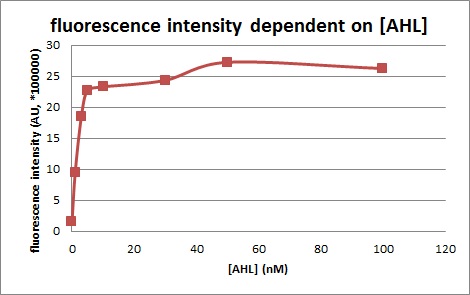
We confirmed fluorescence intensity of LuxR activation promoter is dependent on 3OC6HSL concentration. The AHL concentration which shows half of maximam activity is less than 5nM.
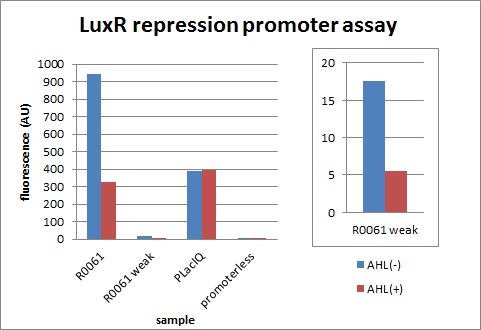
R1, characterization of R0061 & K395008 (promoter repressed by LuxR/3OC6HSL)
R1-1, R0061 (promoter repressed by LuxR/3OC6HSL)
Next, we characterized the existing part R0061, LuxR repression promoter. We examined whether the amount of transcription is appropriate when signal is off and how much this promoter represses.
The expression of GFP with 100nM 3OC6HSL dropped to 1/3 comparing with the expression without 3OC6HSL.
R1-2, K395008 (promoter repressed by LuxR/3OC6HSL):R0061 weak
We confirmed R0061 and found increase of cells was inhibited due to a high level of expression although it is repressed by AHL. Therefore, we designed a new appropriate promoter by changing one base of R0061.
The expression of GFP with 100nM 3OC6HSL dropped to 1/3 comparing with the expression without 3OC6HSL. We found the level of expression is appropriate and this promoter work as expected.
Conclusion
We designed the new promoter which is repressed LuxR/3OC6HSL complex by changing one base of existing promoter. We confirmed this promoter works as we expected.
It is not so difficult to make the promoter which strength is between these two by designing.
Material & Methods
M0, characterization of R0062 (promoter activated by LuxR/3OC6HSL)
fluorescence intensity in the presence/absence of AHL
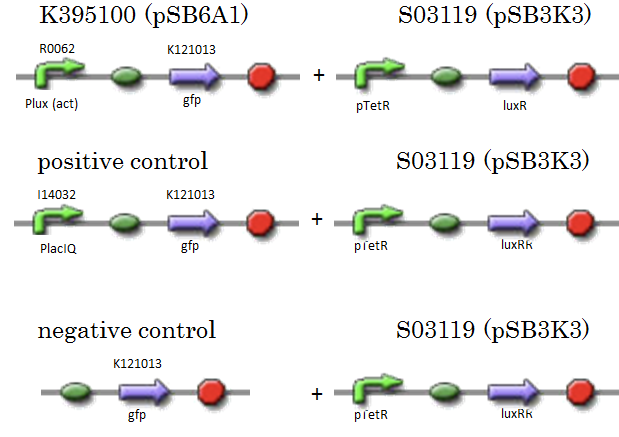
We constructed K395100 combining R0062 and K121013. K121013 is a promoter-less gfp reporter (rbs-gfp-ter-ter) on pSB6A1. S03119 is a LuxR generator which is regulated by PTetR, which is repressed by TetR. In this experiment, we don’t use TetR, so S03119 functions as a LuxR constitutive generator. The backbone of S03119 is pSB1A2, which is a high copy plasmid, so we changed the backbone from pSB1A2 to pSB3K3. We used a fusion of PlacIq (I14032) to gfp (K121013) as a positive control and used promoterless gfp (K121013) as a negative control.
- samples
- [Plux act - gfp](BBa_K395100) on pSB6A1 + [PtetR – LuxR] on pSB3K3
- positive control: [PlacIq(constitutive promoter) - gfp] on pSB6A1+ [PtetR – LuxR] on pSB3K3
- negative control:. [promoterless - gfp] on pSB6A1+ [PtetR – LuxR] on pSB3K3
- Strain
DH5α
- protocol
- Prepare overnight culture.
- Take 30 ul of the overnight culture into LB + antibiotics (Amp + Kan). (→fresh culture)
- Incubate the fresh culture until the observed O.D. reaches around 0.60.
- Each sample was divided into 2. Prepare and add 3OC6HSL mixture to one, and add DMSO mixture to the other. The final concentration of 3OC6HSL is 100nM.
- Induction for 3 hours at 37°C.
- Fluorometer (FLA5200) and flow cytometry measurements for GFP expression.
fluorescence intensity dependent on [AHL]
- samples
- [Plux act - gfp](BBa_K395100) on pSB6A1 + [ptet – LuxR] on pSB3K3
- Strain
DH5α
- protocol
- Prepare overnight culture.
- Take 30 ul of the overnight culture into LB + antibiotics (Amp + Kan). (→fresh culture) Prepare the same 7 tubes for each sample.
- Incubate the fresh culture until the observed O.D. reaches around 0.60.
- Each sample was divided into 2. Prepare and add 3OC6HSL mixture. The final concentration of 3OC6HSL is 1, 3, 5, 10, 30, 50, 100nM.
- Induction for 3 hours at 37°C.
- Fluorometer (FLA5200) and flow cytometry measurements for GFP expression.
M1, characterization of R0061 & K395008 (promoter repressed by LuxR/3OC6HSL)
M1-1, characterization of R0061 (promoter repressed by LuxR/3OC6HSL)
We constructed K395101 combining R0061 and K121013, which is a promoter-less gfp reporter (rbs-gfp-ter-ter) on pSB6A1. S03119 is a LuxR generator which is repressed by TetR. In this experiment, we don’t use TetR, therefore, S03119 functions a LuxR constitutive generator. The backbone of S03119 is pSB1A2, which is a high copy plasmid, so we changed the backbone from pSB1A2 to pSB3K3. We used a fusion of PlacIq (I14032) to gfp (K121013) as a positive control and used promoterless gfp (K121013) as a negative control.
- samples
- [Plux rep - gfp](BBa_K395101) on pSB6A1 + [PtetR – LuxR] on pSB3K3
- positive control: [PlacIq(constitutive promoter) - gfp] on pSB6A1+ [PtetR – LuxR] on pSB3K3
- negative control: [promoterless - gfp] on pSB6A1+ [PtetR – LuxR] on pSB3K3
- Strain
DH5α
- protocol
- Prepare overnight culture.
- Take 30 ul of the overnight culture into LB + antibiotics (Amp + Kan).(→fresh culture)
- Incubate the fresh culture until the observed O.D. reaches around 0.80.
- Each sample was divided into 2. Prepare and add 3OC6HSL mixture to one, and add DMSO mixture to the other. The final concentration of AHL is 100nM.
- Induction for 2 hours at 37°C.
- Fluorometer (FLA5200) and flow cytometry measurements for GFP expression.
M1-2, characterization of K395008 (promoter repressed by LuxR/3OC6HSL):R0061 weak
We constructed K395105 combining K395008 and K121013. K121013 is a promoter-less gfp reporter (rbs-gfp-ter-ter) and this backbone is pSB6A1. Promoter of S03119 is PtetR, which is repressed by tetR. In this experiment, we don’t use TetR, so, S03119 functions a LuxR constitutive generator. The backbone of S03119 is pSB1A2, which is a high copy plasmid, so we changed the backbone from pSB1A2 to pSB3K3. We used a fusion of PlacIq (I14032) to gfp (K121013) as a positive control and used promoterless gfp (K121013) as a negative control.
- samples
- [R0061weak - gfp](BBa_K395105) on pSB6A1 + [PtetR – LuxR] on pSB3K3
- positive control: [PlacIq(constitutive promoter) - gfp] on pSB6A1+ [PtetR – LuxR]) on pSB3K3
- negative control: [promoterless - gfp] on pSB6A1+ [PtetR – LuxR] on pSB3K3
- Strain
DH5α
- protocol
- Prepare overnight culture.
- Take 30 ul of the overnight culture into LB + antibiotics (Amp + Kan). (→fresh culture)
- Incubate the fresh culture until the observed O.D. reaches around 0.60.
- Each sample was divided into 2. Prepare and add 3OC6HSL mixture to one, and add DMSO mixture to the other. The final concentration of 3OC6HSL is 100nM.
- Induction for 3 hours at 37°C.
- Fluorometer (FLA5200) and flow cytometry measurements for GFP expression.
Reference
- KRISTI A. EGLAND & E. P. GREENBERG, Conversion of the Vibrio fischeri Transcriptional Activator LuxR, to a Repressor. JOURNAL OF BACTERIOLOGY, Feb. 2000, p. 805–811
</div>
</div>
 "
"