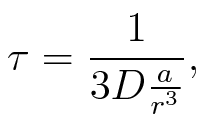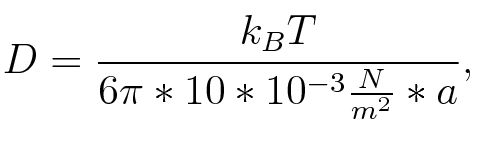Team:TU Munich/Modeling
From 2010.igem.org
|
||||||||||||||||||||
|
|
OverviewWe simulated the termination and antitermination properties of our signal-terminator constructs with the Kinefold web server[1] and used some standard estimations for diffusive terms. Our main goal was to prove that our constructs work and that termination is stopped efficiently, that is, that the signal molecule binds and antiterminations occurs before the RNA polymerase falls off. ResultsDiffusionThe question whether antitermination occurs is not only guided by the folding process of the signal-terminator pair, but also by how long the signal takes to diffuse to the terminator sequence.
To account for the diffusion time, we estimated the hit rate τ (following 6.), which is the time until the signal meets the terminator sequence for the first time:
where D is the diffusion constant, a the radius of gyration of the signal molecule and r the radius of the cell.
where n is the length of the signal which is 0,3 nm/monomer, l is the persistency length which is following (5.) 2nm for single-stranded RNA. Thus, for a signal of length 32 nt, a = 6,4 nm. The diffusion constant D was obtained by
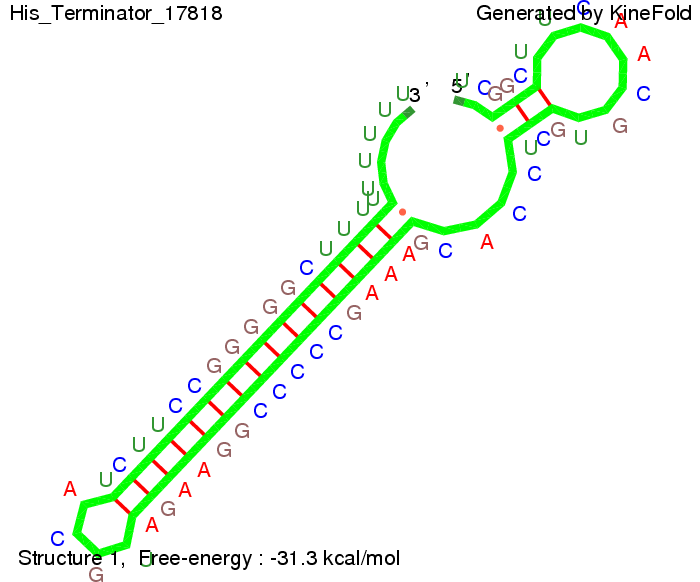
where kB is the Boltzmann constant and T is the absolute temperature. Thus, for a cell containing 100 signal molecules, the signal needs 0,1518 sec until it first hits the terminator sequence. CloseAs the folding time (at least 1,7 sec for full signal length) is significantly larger than the estimated diffusion time (0,1518 sec), diffusion does not play an important role and thus can be neglected in the modeling our signal-terminator constructs. SwitchHis-terminatorTerminator sequence: Both terminator and signal sequence comprise two parts:
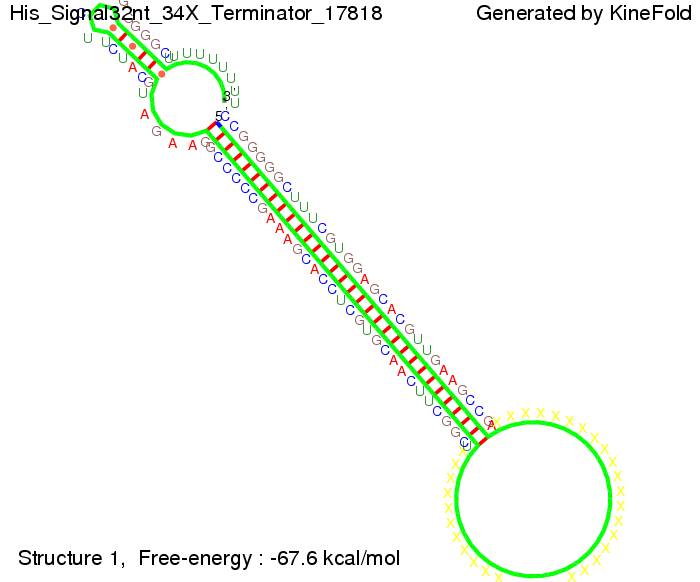
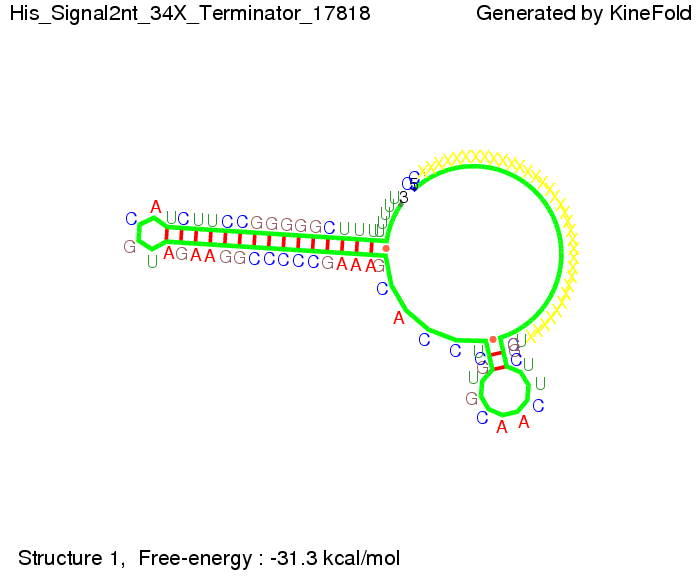
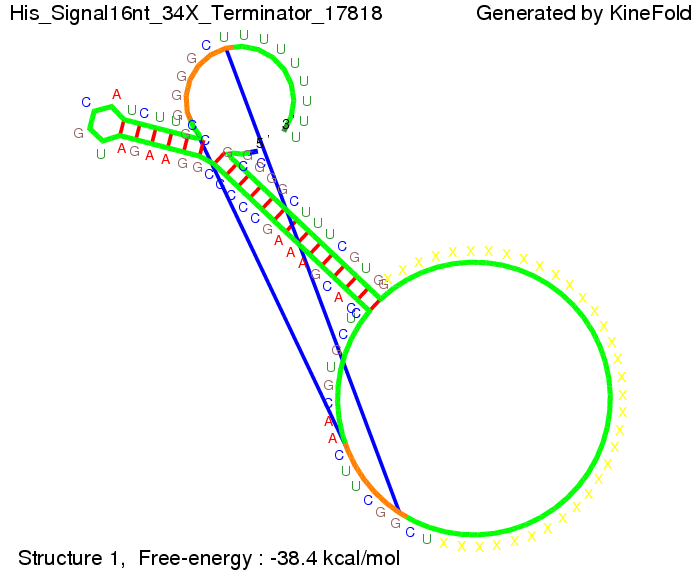
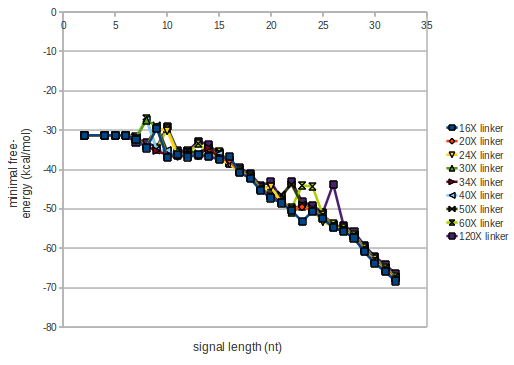
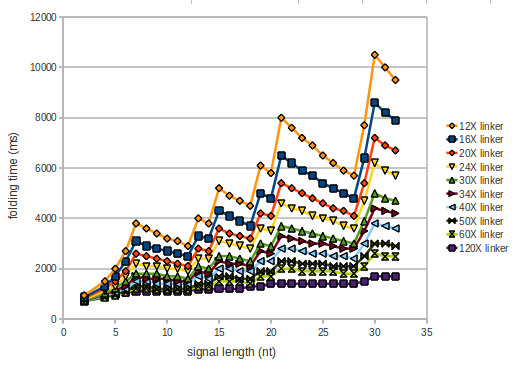
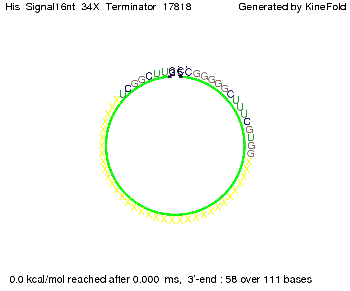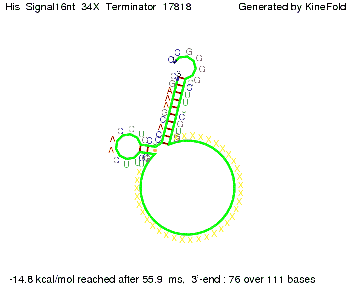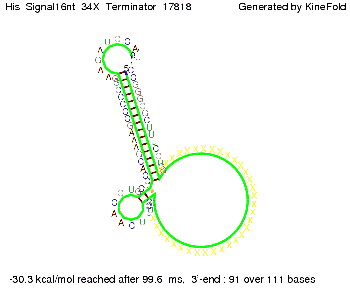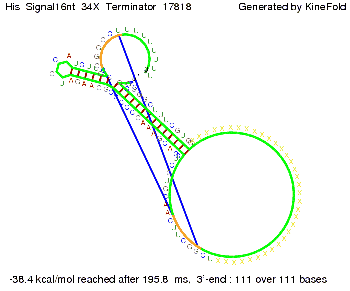
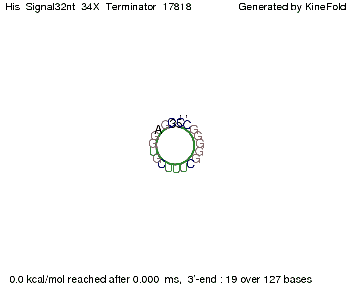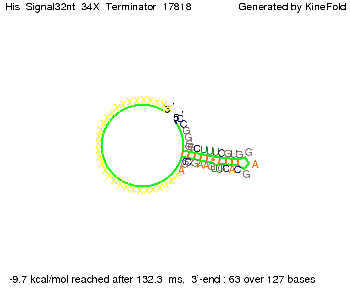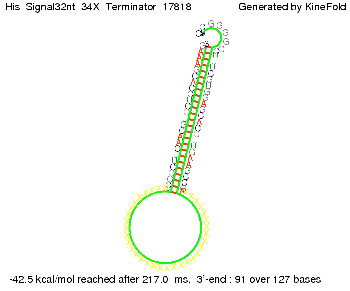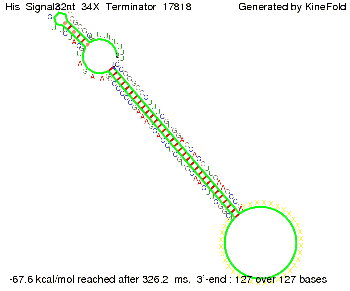
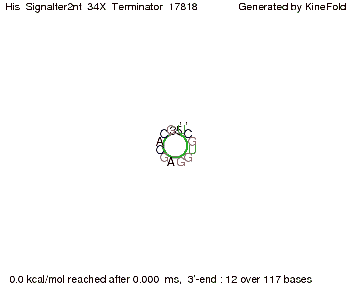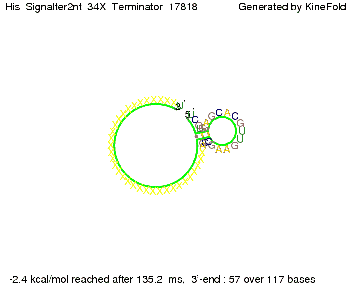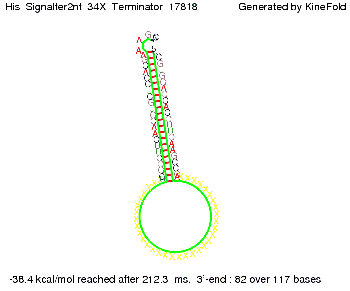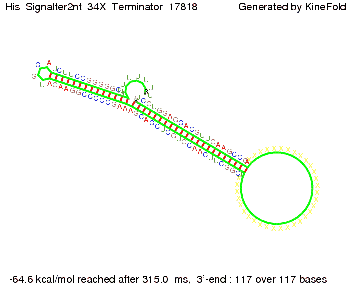
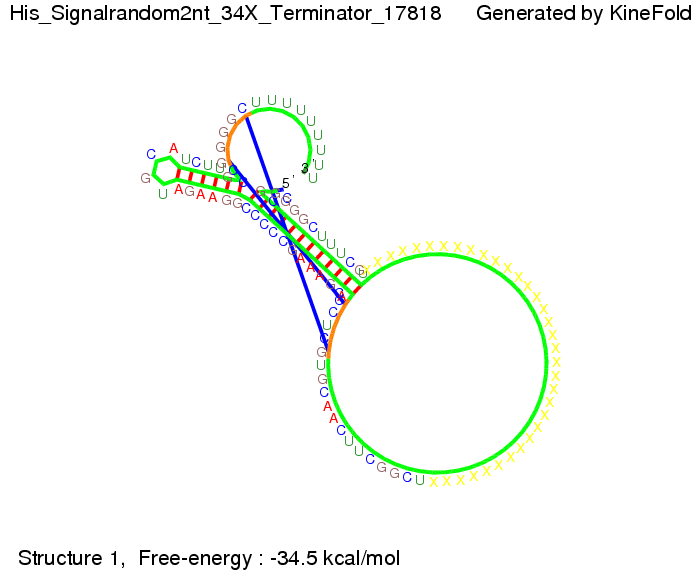
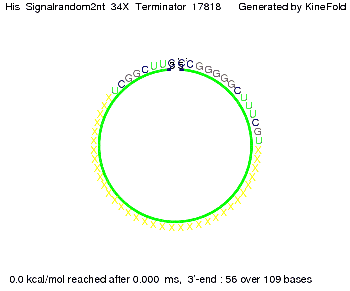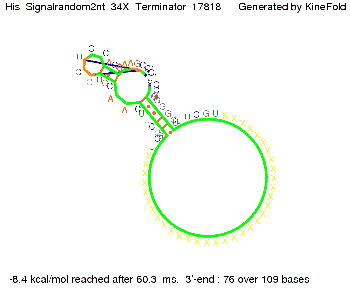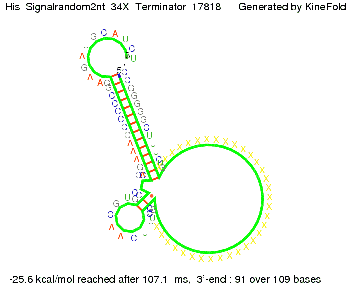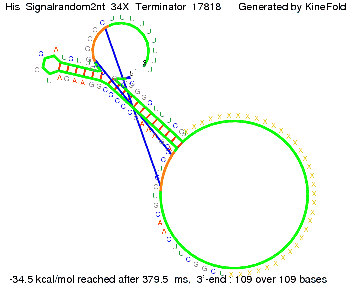
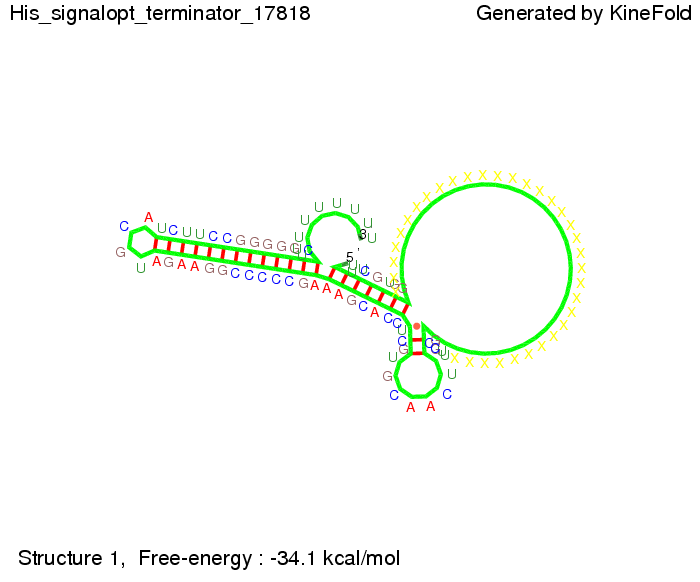
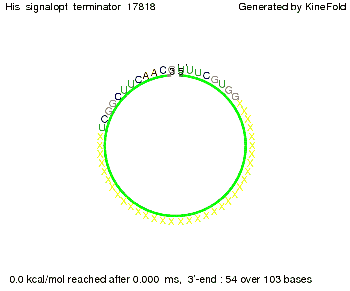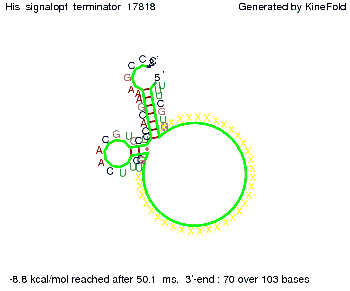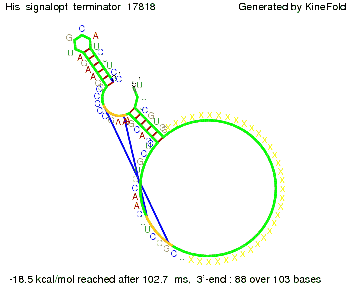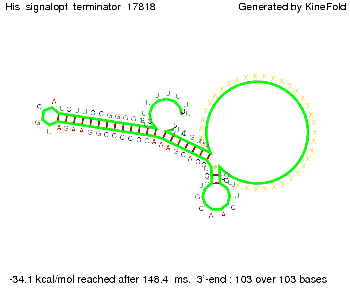
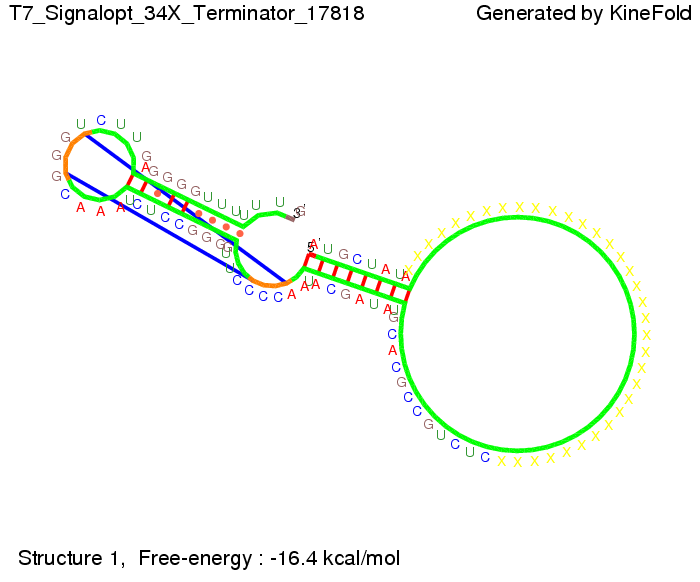
We modeled the signal-terminator interaction for 2, 4-32 nt signal length and proved that the signal sequence binds to the terminator sequence and impedes the terminator folding, thus the polymerase doesn't fall off and the output signal is produced. The minimal free-energy structure of the folded terminator including the random signaling part is shown in the figure "His-terminator" below: Our full signal sequence of length 32 nt completly binds to the terminator and antitermination is successful: On the other hand the picture for a signal length of only 2 nt shows that such a small signal length is not sufficient for antitermination. But already a signal of length 16 nt binds to the terminator so that it cannot fold completely anymore. As we observed a dependence of the folding time and minimal energy respectively from the linker length we used linkers of length 12, 16, 20, 24, 30, 34, 40, 50, 60, 120 'X' bases. For the minimal energy there is only a very small linker length dependence as one can see in the figure below. The figure also shows that the minimal free-energy structure for the signal sequence bound to the terminator sequence is much more stable, around 35 kcal/mol, than the minimal free-energy structure for the terminator sequence alone. However, for the folding time linker length dependence is significant as one can see in the figure below. But, as, for us, the fact whether the terminator is folding or not is crucial and the videos below show that for appropriate signal length the terminator is not folding. Concerning this fact, for all linker lengths we tested there was only a dependence on the signal length but the results were independent from the linker length. Hence, the linker length dependence can be neglected. RNA folding path videosThe video on the left for 16 nt signal length shows that the terminator is already hindered from folding completely and the video on the right, which is done for full signal length of 32 nt, displays that the terminator does not fold at all as the signal immediately binds to the terminator sequence as it is synthesized.
Optimization of the signal sequenceWe also tried to optimize our signal sequence to find the minimal sequence length which still enables antitermination. For this optimization kept the random signaling part constant and varied the part which partially binds to the His-terminator and vice versa to see which length is sufficient for antitermination for each case. Then we combined the results to find a optimal signal.
When varying the part which partially binds to the His-terminator we found that having only the 2 nt of this part together with the random signaling part suffices for antitermination as the picture and the corresponding video below show.
For the full 12 nt of the part which partially binds to the His-terminator, the random signaling part has to have at least 2 nt in order to prevent termination as one can see in the picture below.
We then combined these two results and found that a signal with overall 8 nt, 4 nt for the random signaling part and 4 nt for the part which partially bind to the His-terminator, 5' UUUCGUGG 3', suffice to enable antitermination as the picture and the corresponding video below display.
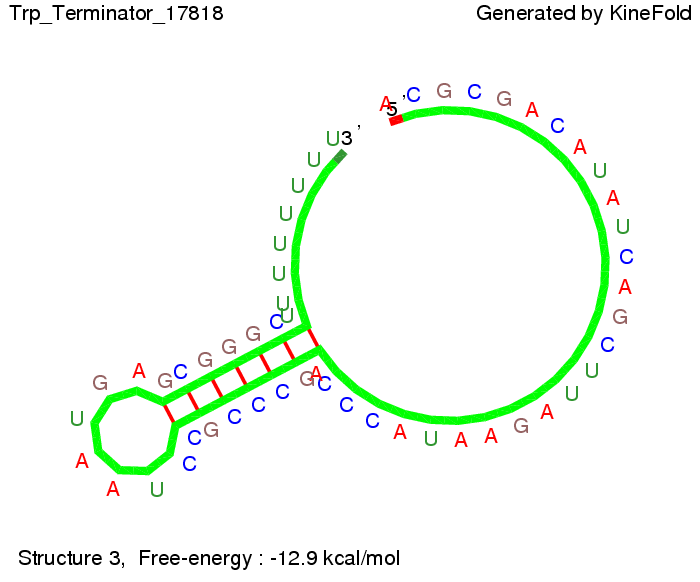
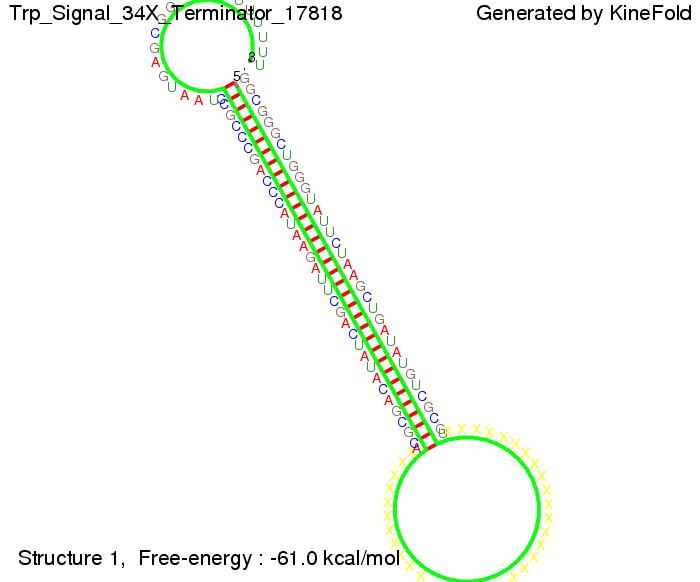
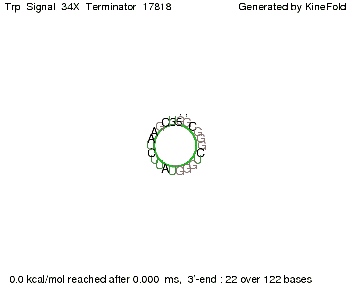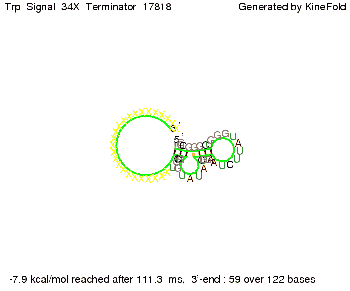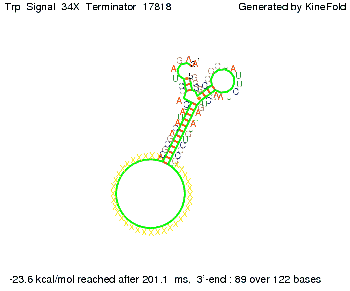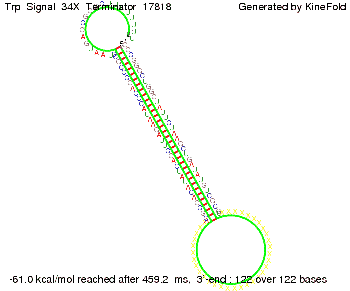
Trp-terminatorTerminator sequence: Both terminator and signal sequence comprise three parts:
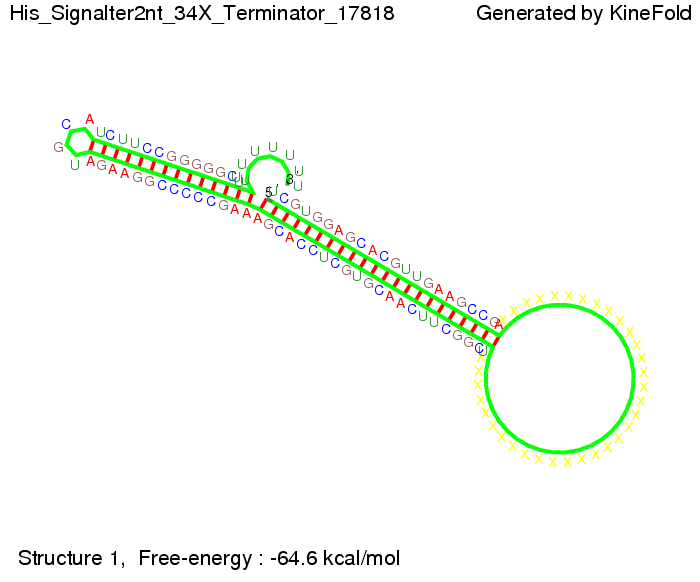
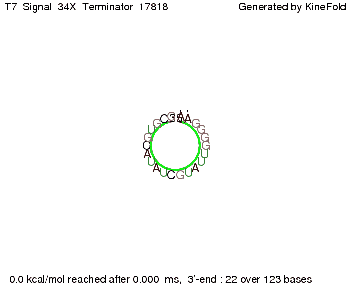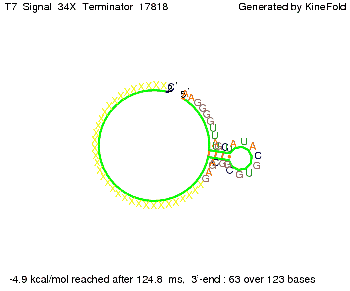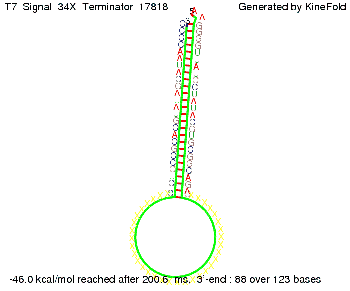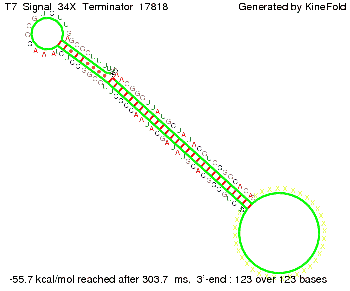
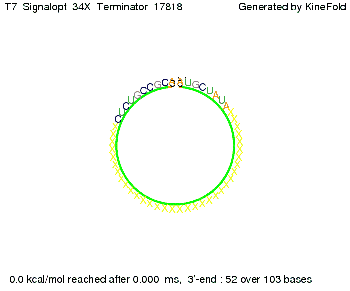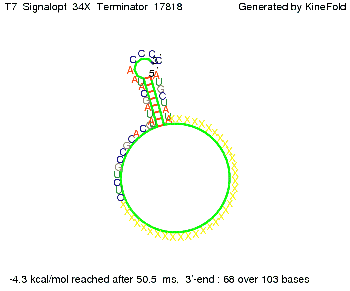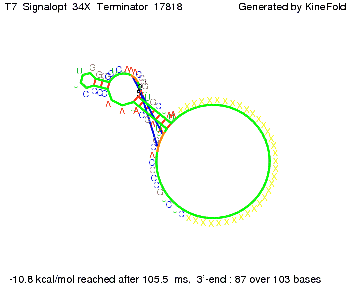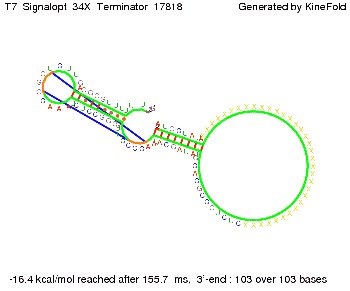
Our full signal sequence of length 34 nt completly binds to the terminator and antitermination is successful:
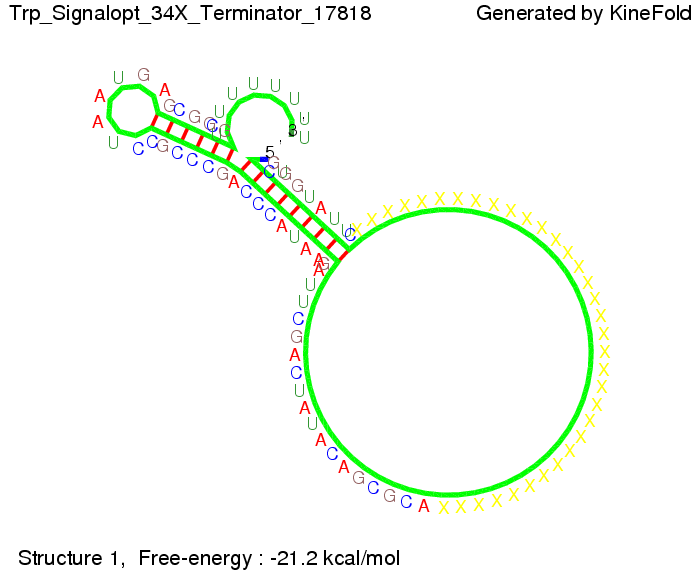
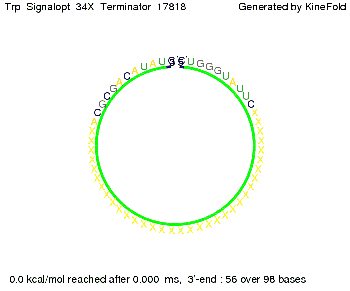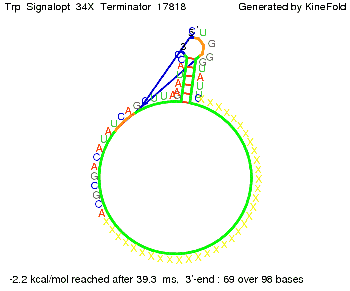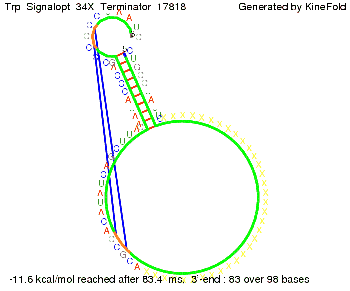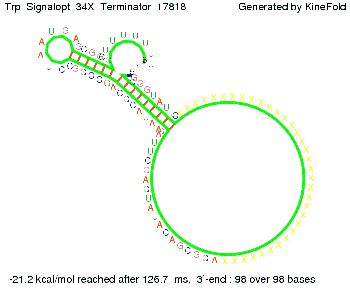
Optimization of the signal sequenceAs for the His-terminator, we tried to optimize our signal sequence to find the minimal sequence length which still enables antitermination. It turned out that a signal with overall 10 nt, 2 nt for the random signaling part, all 4 nt for the part which binds to the upstream sequence of the terminator and 2 nt for the part which partially bind to the Trp-terminator, 5' CUGGGUAUUC 3', suffice to enable antitermination as the picture and the corresponding video below display.
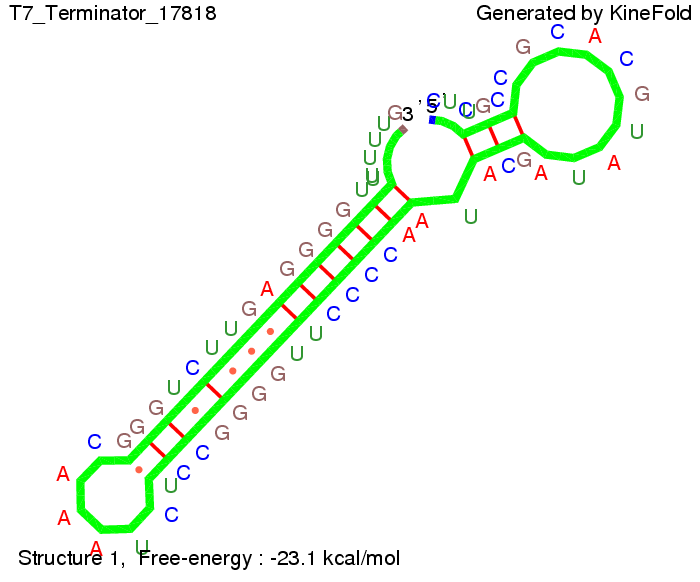
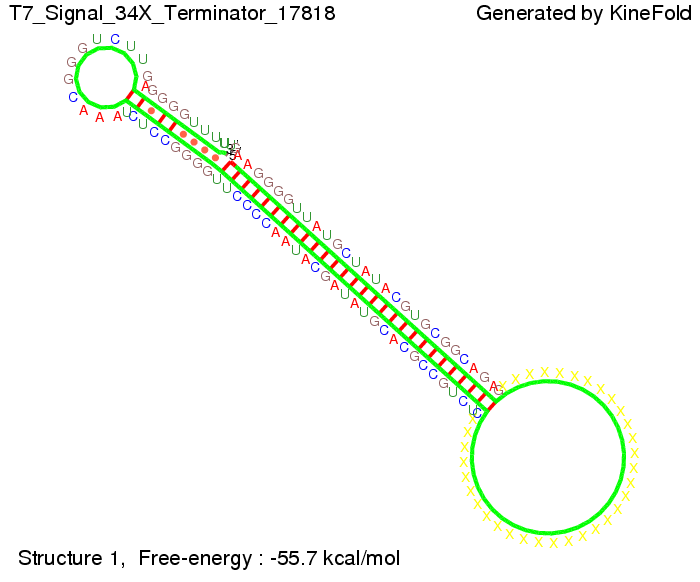
T7-terminatorTerminator sequence: Both terminator and signal sequence comprise two parts:
Our full signal sequence of length 28 nt completly binds to the terminator and anti-termination is successful:
Optimization of the signal sequenceAs for the His-terminator, we tried to optimize our signal sequence to find the minimal sequence length which still enables antitermination. We finaly found that a signal with overall 8 nt, 3 nt for the random signaling part and 5 nt for the part which partially bind to the T7-terminator, 5' AUGCUAUA 3', suffice to enable anti-termination as the picture and the corresponding video below display.
NetworkOutlookIn future our switch could be used to interconnect BioBricks as demonstrated on our [TU Munich/Software| Software] page. Furthermore we have discovered that the currently available RNA folding programs are not fully suitable for describing complex RNA interactions between multiple RNA sequences. This obstacle has to be overcome to allow reliable in silcio modeling of RNA devices in the future. References[1] A. Xayaphoummine, T. Bucher and H. Isambert, Kinefold web server for RNA/DNA folding path and structure prediction including pseudoknots and knots, 2005, Nucleic Acids Research, Vol. 33, Web Server issue W605–W610. [2] A. Xayaphoummine, T. Bucher, F. Thalmann, and H. Isambert, Prediction and statistics of pseudoknots in RNA structures using exactly clustered stochastic simulations, December 23, 2003, 15310–15315 PNAS vol. 100 no. 26. [3] Christoph Flamm, Walter Fontana, Ivo L. Hofacker, and Peter Schuster, RNA folding at elementary step resolution, RNA (2000), 6:325–338 Cambridge University Press. [4] David H. Mathews, Jeffrey Sabina, Michael Zuker and Douglas H. Turner, Expanded Sequence Dependence of Thermodynamic Parameters Improves Prediction of RNA Secondary Structure, 1999, J. Mol. Biol. (1999) 288, 911-940. [5] Rippe, Making contacts on a nucleic acid polymer, 2001, TRENDS in Biochemical Sciences. [6] Berg, v. Hippel, 1985. |
|||||||||||||||||||
 "
"