Team:TU Delft/Project/sensing/results
From 2010.igem.org
(→BBa_K398326) |
(→pCaiF strength) |
||
| Line 6: | Line 6: | ||
===pCaiF strength=== | ===pCaiF strength=== | ||
====[http://partsregistry.org/Part:BBa_K398326 BBa_K398326]==== | ====[http://partsregistry.org/Part:BBa_K398326 BBa_K398326]==== | ||
| - | pCaiF is one of our Biobricks of the sensing project. It's function is to express proteins under glucose limitation conditions. In order to measure its strength we attached the GFP generator [http://partsregistry.org/Part:BBa_E0240 BBa_E0240] in front of this new promoter in order to know the response produced in the different growth phases. | + | [http://partsregistry.org/Part:BBa_K398326 BBa_K398326 (pCaiF)] is by far the smallest part of our project; as we explained before we looked for a promoter that could enable the expression proteins under low glucose concentrations in order to mimic a diauxic shift for the alkane degradation system: once the glucose starts to be limiting factor the expression of alkane degradation genes under pCaiF control will enable (theoreticallly) to the cells shift from glucose metabolism to alkane degradation, everything done by a piece of just 51 bp. |
| + | |||
| + | The pCaiF regulation mechanism is really simple, [http://partsregistry.org/Part:BBa_K398326 BBa_K398326 (pCaiF)] contains a cAMP-crp complex binding domain, cAMP-crp is known as transcriptional regulator. When glucose concentrations in the surroundings are high, cAMP levels are low because there is a lot of energy source that can be metabolized by the cells; however during starvation periods cAMP levels increase and thus the concentration of the complex cAMP-crp activating at least 180 genes related to starvation response. Among those CaiF a protein used during Carnitine anaerobic metabolism. | ||
| + | |||
| + | Due to time constraints we couldn't make a more elegant circuit or express our proteins using [http://partsregistry.org/Part:BBa_K398326 BBa_K398326 (pCaiF)], however we wanted to show that the part does what we expected: Protein production at low glucose concentrations. We attached a GFP generator in order to measure the output of our part. First we used a rich medium (LB) and we diluted it with M9 medium without glucose (50% v/v) in order to show the differences in protein expression when the carbon source concentration is lower in a rich medium. | ||
| + | |||
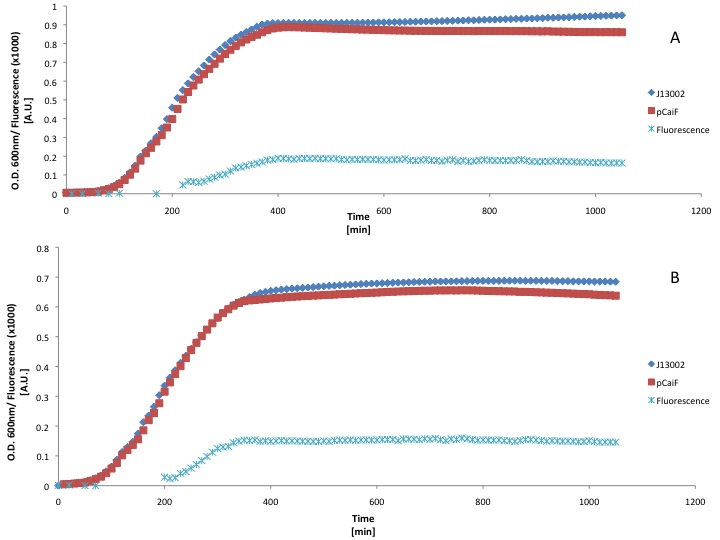
| + | [[Image:TU_Delft_pCaiF_LB.jpg|thumb|center| GFP and Biomass profiles in LB growth. (A) Normal LB and (B) LB diluted 50% with M9 medium. ]] | ||
| + | |||
| + | Our findings suggest that there is a slight improvement on GFP production when the LB medium is diluted 50%. However the fluorescence produced is a really low signal and we didn't see an increase of GFP production during the stationary phase (starvation). This could be due to the fact that LB medium can keep the cAMP levels low for long periods of time. | ||
| + | |||
| + | |||
| + | one of our Biobricks of the sensing project. It's function is to express proteins under glucose limitation conditions. In order to measure its strength we attached the GFP generator [http://partsregistry.org/Part:BBa_E0240 BBa_E0240] in front of this new promoter in order to know the response produced in the different growth phases. | ||
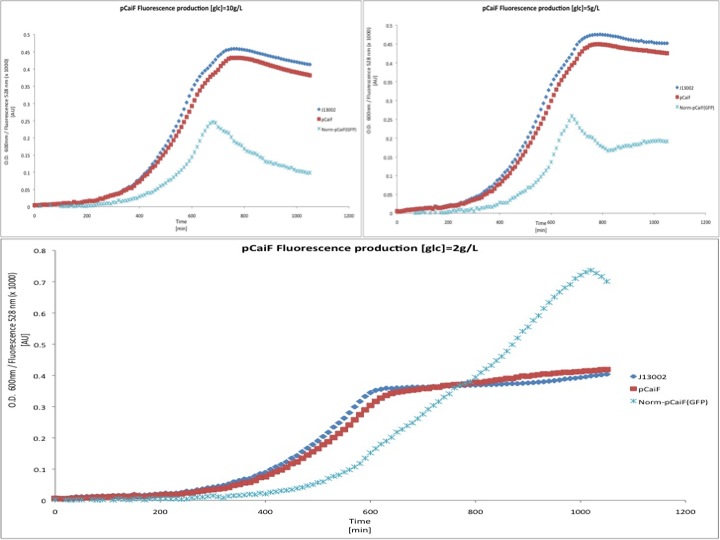
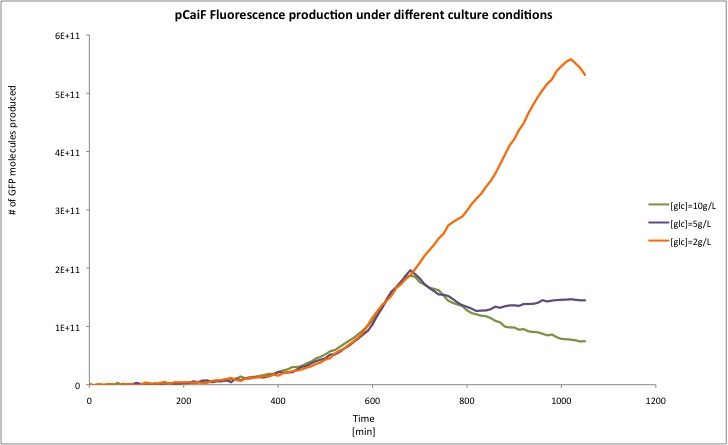
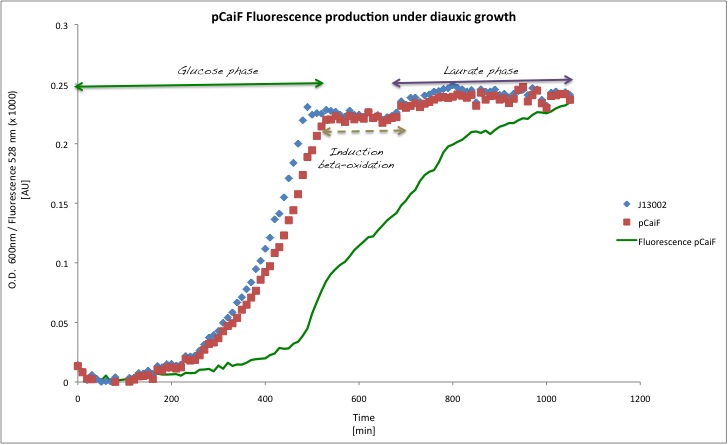
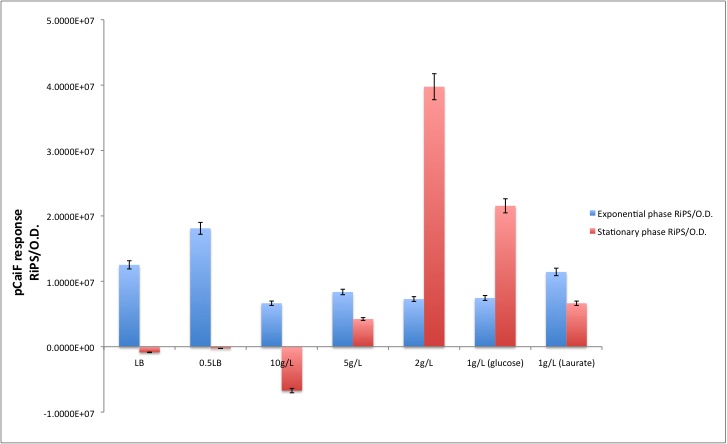
Our results are shown in the following plots: | Our results are shown in the following plots: | ||
| - | |||
[[Image:TU_Delft_pCaiF_glucose.jpg]] | [[Image:TU_Delft_pCaiF_glucose.jpg]] | ||
Revision as of 11:31, 27 October 2010

Sensing Results
pCaiF strength
BBa_K398326
BBa_K398326 (pCaiF) is by far the smallest part of our project; as we explained before we looked for a promoter that could enable the expression proteins under low glucose concentrations in order to mimic a diauxic shift for the alkane degradation system: once the glucose starts to be limiting factor the expression of alkane degradation genes under pCaiF control will enable (theoreticallly) to the cells shift from glucose metabolism to alkane degradation, everything done by a piece of just 51 bp.
The pCaiF regulation mechanism is really simple, BBa_K398326 (pCaiF) contains a cAMP-crp complex binding domain, cAMP-crp is known as transcriptional regulator. When glucose concentrations in the surroundings are high, cAMP levels are low because there is a lot of energy source that can be metabolized by the cells; however during starvation periods cAMP levels increase and thus the concentration of the complex cAMP-crp activating at least 180 genes related to starvation response. Among those CaiF a protein used during Carnitine anaerobic metabolism.
Due to time constraints we couldn't make a more elegant circuit or express our proteins using BBa_K398326 (pCaiF), however we wanted to show that the part does what we expected: Protein production at low glucose concentrations. We attached a GFP generator in order to measure the output of our part. First we used a rich medium (LB) and we diluted it with M9 medium without glucose (50% v/v) in order to show the differences in protein expression when the carbon source concentration is lower in a rich medium.
Our findings suggest that there is a slight improvement on GFP production when the LB medium is diluted 50%. However the fluorescence produced is a really low signal and we didn't see an increase of GFP production during the stationary phase (starvation). This could be due to the fact that LB medium can keep the cAMP levels low for long periods of time.
one of our Biobricks of the sensing project. It's function is to express proteins under glucose limitation conditions. In order to measure its strength we attached the GFP generator BBa_E0240 in front of this new promoter in order to know the response produced in the different growth phases.
Our results are shown in the following plots:
| Condition | Exponential phase [RiPS/O.D.] | Stationary phase [RiPS/O.D.] |
|---|---|---|
| LB | 1.2516E+07 | -8.6245E+05 |
| 0.5LB | 1.8101E+07 | -2.5945E+05 |
| 10g/L | 6.6456E+06 | -6.7109E+06 |
| 5g/L | 8.3673E+06 | 4.2339E+06 |
| 2g/L | 7.2869E+06 | 3.9755E+07 |
| 1g/L (glucose) | 7.4517E+06 | 2.1543E+07 |
| 1g/L (Laurate) | 1.1435E+07 | 6.6428E+06 |

 "
"