Team:TU Delft/Project/rbs-characterization/parts
From 2010.igem.org
(→Experimental Setup) |
(→BioBricks, the making of the RBS Characterization) |
||
| Line 5: | Line 5: | ||
==BioBricks, the making of the RBS Characterization== | ==BioBricks, the making of the RBS Characterization== | ||
| + | |||
| + | ===Experimental setup=== | ||
| + | The following RBS sequences from the [http://partsregistry.org/Ribosome_Binding_Sites/Prokaryotic/Constitutive/Anderson Anderson RBS family] were used: | ||
| + | *[http://partsregistry.org/Part:BBa_J61100 BBa_J61100] | ||
| + | *[http://partsregistry.org/Part:BBa_J61101 BBa_J61101] | ||
| + | *[http://partsregistry.org/Part:BBa_J61107 BBa_J61107] | ||
| + | *[http://partsregistry.org/Part:BBa_J61117 BBa_J61117] | ||
| + | *[http://partsregistry.org/Part:BBa_J61127 BBa_J61127] | ||
| + | |||
| + | For the purpose of comparison and standardization the following RBS was used as a reference for our characterization. | ||
| + | *[http://partsregistry.org/Part:BBa_B0032 BBa_B0032] | ||
| + | |||
| + | These RBS sequences were positioned upstream of the standard [http://partsregistry.org/Part:BBa_E0040 GFP coding sequence], so expression could be measured. | ||
BBa_I13401 was PCR amplified using the universal primers G00100 and G00101. The purified PCR product was assembled into one of the Anderson RBS plasmids provided in the distribution plates by means of 2-way ligations. | BBa_I13401 was PCR amplified using the universal primers G00100 and G00101. The purified PCR product was assembled into one of the Anderson RBS plasmids provided in the distribution plates by means of 2-way ligations. | ||
| Line 10: | Line 23: | ||
Transformation into competent Top10 E.coli cells yielded positives, as determined by fluorescence analysis. | Transformation into competent Top10 E.coli cells yielded positives, as determined by fluorescence analysis. | ||
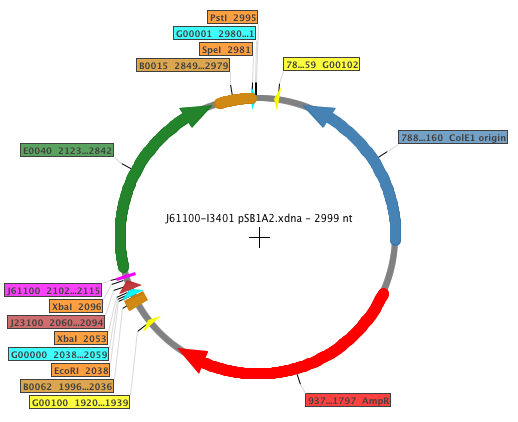
| - | The | + | The Biobricks generated in order to perform the experiments were: [http://partsregistry.org/Part:BBa_K398500 K398500], [http://partsregistry.org/Part:BBa_K398501 K398501], [http://partsregistry.org/Part:BBa_K398502 K398502], [http://partsregistry.org/Part:BBa_K398503 K398503], [http://partsregistry.org/Part:BBa_K398504 K398504]. The general map of the construction is shown below, where the RBS is displayed in fucsia. |
| + | |||
| + | [[Image:RBS1.jpg|left|650px]] | ||
| + | |||
| + | {| style="color:black; background-color:white;" cellpadding="5" cellspacing="0" border="1" | ||
| + | |'''Feature''' | ||
| + | |'''Function''' | ||
| + | |- | ||
| + | |[http://partsregistry.org/Part:BBa_B0032 AmpR] | ||
| + | |Ampicillin resistance | ||
| + | |- | ||
| + | |[http://partsregistry.org/Part:BBa_B0015 B0015] | ||
| + | |Transcriptional (double) terminator | ||
| + | |- | ||
| + | |[http://partsregistry.org/Part:BBa_B0062 B0062] | ||
| + | |Transcriptional terminator | ||
| + | |- | ||
| + | |[http://partsregistry.org/Part:BBa_E0040 E0040] | ||
| + | |GFP | ||
| + | |- | ||
| + | |[http://partsregistry.org/Part:BBa_G00000 G00000] | ||
| + | |Standard prefix | ||
| + | |- | ||
| + | |[http://partsregistry.org/Part:BBa_G00001 G00001] | ||
| + | |Standard suffix | ||
| + | |- | ||
| + | |[http://partsregistry.org/Part:BBa_G00100 G00100] | ||
| + | |VF2 primer binding site | ||
| + | |- | ||
| + | |[http://partsregistry.org/Part:BBa_G00101 G00101] | ||
| + | |VR primer binding site | ||
| + | |- | ||
| + | |[http://partsregistry.org/Part:BBa_J61100 J61100] | ||
| + | |RBS Anderson family | ||
| + | |- | ||
| + | |[http://partsregistry.org/Part:BBa_J23100 J23100] | ||
| + | |Promoter | ||
| + | |} | ||
| + | |||
| + | <html><div style='clear:both'> | ||
| + | </html> | ||
== Parts == | == Parts == | ||
Revision as of 14:53, 23 October 2010

BioBricks, the making of the RBS Characterization
Experimental setup
The following RBS sequences from the Anderson RBS family were used:
For the purpose of comparison and standardization the following RBS was used as a reference for our characterization.
These RBS sequences were positioned upstream of the standard GFP coding sequence, so expression could be measured.
BBa_I13401 was PCR amplified using the universal primers G00100 and G00101. The purified PCR product was assembled into one of the Anderson RBS plasmids provided in the distribution plates by means of 2-way ligations.
Transformation into competent Top10 E.coli cells yielded positives, as determined by fluorescence analysis.
The Biobricks generated in order to perform the experiments were: K398500, K398501, K398502, K398503, K398504. The general map of the construction is shown below, where the RBS is displayed in fucsia.
| Feature | Function |
| AmpR | Ampicillin resistance |
| B0015 | Transcriptional (double) terminator |
| B0062 | Transcriptional terminator |
| E0040 | GFP |
| G00000 | Standard prefix |
| G00001 | Standard suffix |
| G00100 | VF2 primer binding site |
| G00101 | VR primer binding site |
| J61100 | RBS Anderson family |
| J23100 | Promoter |
Parts
Our constructs used for measurements: <partinfo>K398500 SpecifiedComponents</partinfo>
<partinfo>K398501 SpecifiedComponents</partinfo>
<partinfo>K398502 SpecifiedComponents</partinfo>
<partinfo>K398503 SpecifiedComponents</partinfo>
<partinfo>K398504 SpecifiedComponents</partinfo>
 "
"