Team:TU Delft/Project/alkane-degradation/parts
From 2010.igem.org
(→BioBricks, the making of) |
(→BioBricks, the making of) |
||
| Line 17: | Line 17: | ||
[[Image:TUDelft_BBa_K398022.png|300px]] | [[Image:TUDelft_BBa_K398022.png|300px]] | ||
| + | |||
| + | =Characterization= | ||
| + | The following strains will be characterized: | ||
| + | • E.coli K12 containing BBa_K398028 in pSB1A2 (AlkB and RubA3) and BBa_K398011 in pSB1C3 (RubR and RubA4) | ||
| + | • E.coli K12 containing BBa_K398017 in pSB1A2 (LadA) | ||
| + | • E.coli K12 containing BBa_K398026 in pSB1A2 (ADH and ALDH) | ||
| + | • E.coli K12 containing BBa_K398026 in pSB1A2 (ADH and ALDH) and BBa_K398017 in pSB1C3 (LadA) | ||
| + | • E.coli K12 containing an ‘empty’ pSB1A2 plasmid | ||
| + | |||
| + | The characterization will involve three main aspects: | ||
| + | 1. The alkane lengths that are converted by the respective genes | ||
| + | 2. The speed at which the alkanes are converted by the enzymes | ||
| + | 3. Possible growth of E.coli K12 on the respective alkane | ||
| + | |||
| + | ===Characterization of growth=== | ||
| + | The preliminary characterization will aim to determine the presence of growth on any one of the following alkanes: | ||
| + | • octane (C8) | ||
| + | • undecane (C11) | ||
| + | • dodecane (C12) | ||
| + | • tetradecane (C14) | ||
| + | • heptadecane (C¬17) | ||
| + | • octadecane (C18) | ||
| + | |||
| + | For the growth characterization on short-chain and long-chain alkanes an E.coli K12 strain containing BBa_K398015 and BBa_K398022 (or intermediates thereof) is inoculated into M9 minimal medium containing 5% v/v ratio of the respective alkane on a 96-well plate with 200 μL volume per well. Growth will be determined o/n by absorbance at 600nm with intervals of 10 minutes. | ||
| + | |||
| + | ===Characterization of enzyme functionality=== | ||
| + | Parallel to this, resting-cell assays will be performed on growth-inhibited E.coli K12 strains containing the constructs described earlier. These assays will indicate the presence or absence of the desired enzymes, regardless of the alkane’s utilization for growth. The hydrocarbon compositions will be determined by gas chromatography analysis. | ||
===''References''=== | ===''References''=== | ||
Revision as of 11:43, 10 September 2010
Contents |
BioBricks, the making of
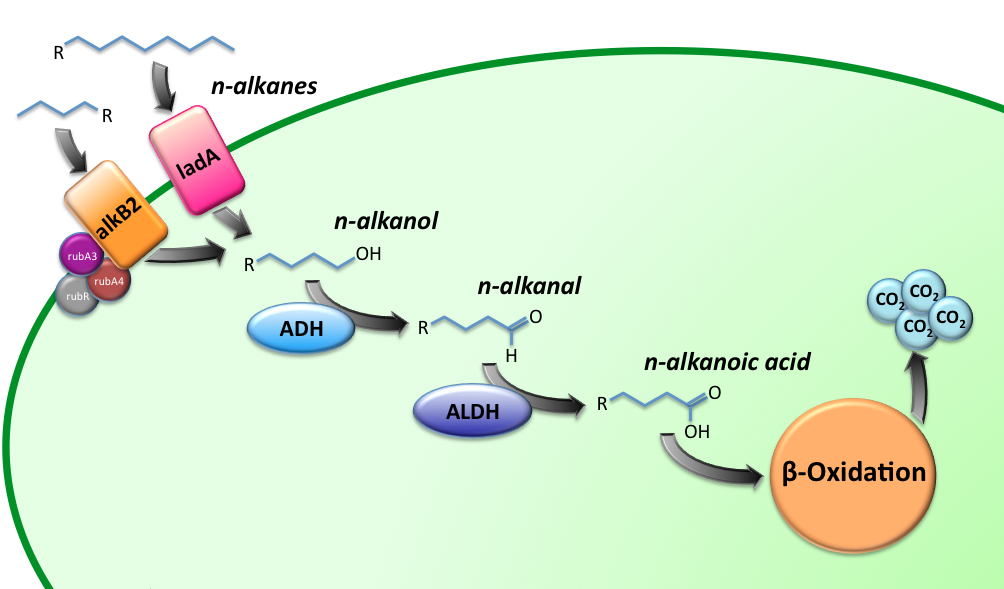
Two BioBricks are the final constructs for this sub-project. The BioBricks K398015 and K398022, implemented in E.coli, will be able to degrade medium (C5-13) and long chain (C15-36) alkanes respectively, while utilizing them as a C-source. The oxidation route of the alkanes is illustrated in figure 1.
To create the alkane degradation constructs a number of genes encoding for alkane degradation enzymes were synthesized and combined with promoters and RBSs obtained from the BioBrick distribution plates. Combination of these genes resulted in the following final BioBrick constructs (the intermediate have also been submitted to the registry).
BBa_K398014 - Short-chain (C5-13) Alkane degradation
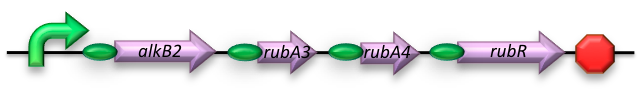
The short-chain hydrocarbon degrading BioBrick contains the AlkB gene cluster from ‘’Gordonia sp. TF6’’[1]. This will facilitate the initial step of the oxidation of C5-13 alkanes as well as that of C5-8 cycloalkanes. It is expected that the in-house mechanism of E.coli will be able to further degrade the products of this pathway. The gene cluster is formed by the genes for alkB2 (alkane 1-monooxygenase), rubA3 (rubredoxin), rubA4 (rubredoxin) and rubR (rubredoxin reductase). alkB2 is a non-haem diiron monooxygenase membrane protein, reported for several genus and species. This monooxygenase oxidizes n-alkanes to the respective n-alkanols and requires three soluble electron-transfer proteins, rubredoxin (rubA3 & rubA4) and rubredoxin reductase (rubR).
BBa_K398022 - Long-chain (C15-36) Alkane degradation
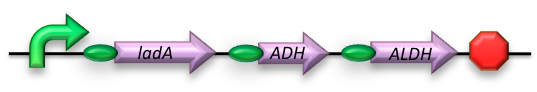
The long-chain degrading BioBrick contains three genes, forming a degradation pathway form alkanes to alkanoic acids. For the first step in this pathway ladA will be used[2], a long-chain alkane monooxygenase from ‘’Geobacillus thermodenitrificans’’ NG80-2. This gene facilitates the conversion of long-chain alkanes (up to at least C36) to their corresponding primary alcohols. Because of the length of the alkanes (and thus also of the pathway intermediates) the BioBrick will also contain an additional ADH and ALDH Bacillus thermoleovorans B23[3-4]. These genes will facilitate the conversion of these long-chain intermediates (alkanols & alkanals).
Characterization
The following strains will be characterized: • E.coli K12 containing BBa_K398028 in pSB1A2 (AlkB and RubA3) and BBa_K398011 in pSB1C3 (RubR and RubA4) • E.coli K12 containing BBa_K398017 in pSB1A2 (LadA) • E.coli K12 containing BBa_K398026 in pSB1A2 (ADH and ALDH) • E.coli K12 containing BBa_K398026 in pSB1A2 (ADH and ALDH) and BBa_K398017 in pSB1C3 (LadA) • E.coli K12 containing an ‘empty’ pSB1A2 plasmid
The characterization will involve three main aspects: 1. The alkane lengths that are converted by the respective genes 2. The speed at which the alkanes are converted by the enzymes 3. Possible growth of E.coli K12 on the respective alkane
Characterization of growth
The preliminary characterization will aim to determine the presence of growth on any one of the following alkanes: • octane (C8) • undecane (C11) • dodecane (C12) • tetradecane (C14) • heptadecane (C¬17) • octadecane (C18)
For the growth characterization on short-chain and long-chain alkanes an E.coli K12 strain containing BBa_K398015 and BBa_K398022 (or intermediates thereof) is inoculated into M9 minimal medium containing 5% v/v ratio of the respective alkane on a 96-well plate with 200 μL volume per well. Growth will be determined o/n by absorbance at 600nm with intervals of 10 minutes.
Characterization of enzyme functionality
Parallel to this, resting-cell assays will be performed on growth-inhibited E.coli K12 strains containing the constructs described earlier. These assays will indicate the presence or absence of the desired enzymes, regardless of the alkane’s utilization for growth. The hydrocarbon compositions will be determined by gas chromatography analysis.
References
[1] Fujii, T., Narikawa, T., Takeda, K., Kato, J., Biotransformation of various alkanes using the Escherichia coli expressing an alkane hydroxylase system from Gordonia sp. TF6. Bioscience, biotechnology, and biochemistry, 68(10) 2171-2177 (2004)
[2] Liu Li, Xueqian Liu, Wen Yang, Feng Xu, Wei Wang, Lu Feng, Mark Bartlam, Lei Wang and Zihe Rao. Crystal Structure of Long-Chain Alkane Monooxygenase (LadA) in Complex with Coenzyme FMN: Unveiling the Long-Chain Alkane Hydroxylase. Journal of molecular biology, 376: 453–465 (2008)
[3] Tomohisa Kato, Asuka Miyanaga, Mitsuru Haruki, Tadayuki Imanaka, Masaaki Morikawa & Shigenori Kanaya. Gene Cloning of an Alcohol Dehydrogenase from Thermophilic Alkane-Degrading Bacillus thermoleovorans B23. Journal of Bioscience and Bioengineering 91(1):100-102 (2001)
[4] Tomohisa Kato, Asuka Miyanaga, Shigenori Kanaya, Masaaki Morikawa. Gene cloning and characterization of an aldehyde dehydrogenase from long-chain alkane-degrading Geobacillus thermoleovorans B23. Extremophiles 14:33-39 (2010)
 "
"