Team:TU Delft/Modeling/interaction-mapping
From 2010.igem.org
Homolog Interaction Mapping (HIM)
Part of our project was development of an application that can generate a list of putative interactions for genes in a new host organism. In our project, the clearest example of this is the insertion of a prefoldin (phPFDα + phPFDβ) coding plasmid into E. coli. Using this application it was possible to get a list of putative interactions with the prefoldin proteins, and therefore give some hints about how these proteins are actually working.
Problems with introducing new genes
In our project, we are taking genes from various organisms and putting them as a biobrick plasmid into E. coli. This may cause various unexpected problems in the newly designed biological system:
- The protein in the original organism may need some other functional partners (proteins or RNAs) in order to behave correctly (I.A.W. a part of the imported system is missing)
- The introduced protein can interact with local proteins that are not ment to interact.
Application
An application was developed that can run these queries on a PostgreSQL server running the STRING database. This database can be downloaded for free for academic use. Unfortunately, the STRING API does not provide enough functionality to do the homolog mapping, so a PostgreSQL database with the STRING data is always needed.
More detailed use of the application is described in the application page.
Searching for interactions
To get an idea of which interactions may occur, interactions within the original organisms where mapped to their E. coli homologs:
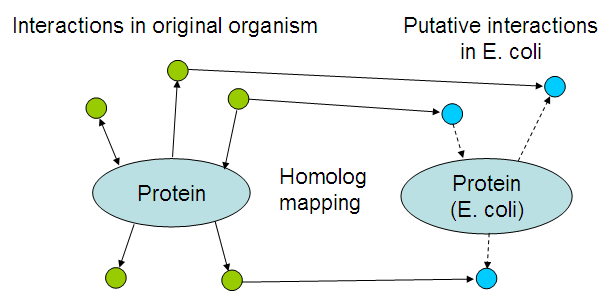
In more detail:
- The sequence of every introduced protein (See the coding biobricks) was blasted against the STRING protein database. This resulted in a number of proteins that are either the correct proteins, or homologs of the introduced proteins.
| Biobrick | Short name | STRING Database ID |
| BBa_K398000 | LadA | 420246.GTNG_3499 |
| BBa_K398001 | AlkB | 101510.RHA1_ro02534 |
| BBa_K398002 | RubA3 | 246196.MSMEG_1840 |
| BBa_K398003 | RubA4 | 350058.Mvan_1744 |
| BBa_K398004 | Rubredoxin reductase | 101510.RHA1_ro02537 |
| BBa_K398005 | Bt-ADH | 235909.GK0984 |
| BBa_K398006 | Bt-ALDH | 235909.GK2772 |
| BBa_K398100 | bbc1 | Dropped: Not in STRING database, and
protein sequence blast only resulted in eukaryotic homologs |
| BBa_K398200 | AlnA | 62977.ACIAD0697 |
| BBa_K398201 | OprG | 160488.PP_0504 |
| BBa_K398300 | AlkS | 399739.Pmen_0429 |
| BBa_K398400 | Prefoldin-alpha | 70601.PH0527 |
| BBa_K398401 | Prefoldin-beta | 70601.PH0532 |
- For each protein, the interactions stored in STRING where gathered.
- For each found interacting protein, E. coli homologs where gathered.
- This results in a combination of interactions with their E. coli homologs. The likelyness of this being a true interaction depends on the STRING interaction score (Data from publication text mining, microarrays and other sources) and the BLAST score (The homolog mapping step).
Result
Prefoldin interaction mapping
The network below shows the putative interactions of the prefoldin biobricks, and any interacting proteins that do not have homologs in E. coli.
- Green: E. coli homologs of a Pyrococcus protein that is interacting with prefoldin, according to the STRING database.
- Red: Missing homolog.
As prefoldin is working in E. coli, one could argue that this is because it is interacting with the green proteins. This could be a hint for more research into the Pyrococcus prefoldin proteins.
Alkane degradation pathway interaction mapping
Additionally, the alkane degradation proteins where also entered into the mapping tool, as shown below. However, we already know how this system works. The STRING interactions probably just show parts of the pathway proteins that are expressed together, hence this analysis is a lot less useful.
 "
"