Team:TU Delft/Modeling/HC regulation/Sensitivity
From 2010.igem.org
Sensitivity analysis
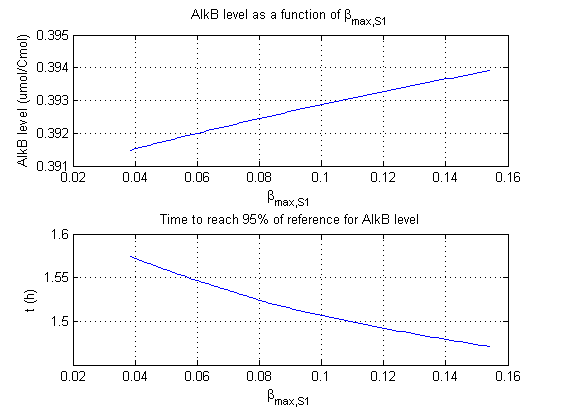
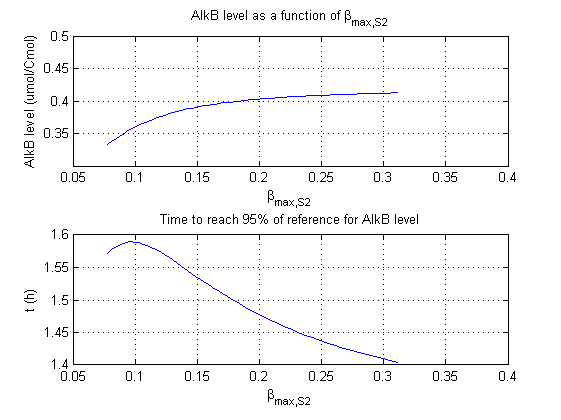
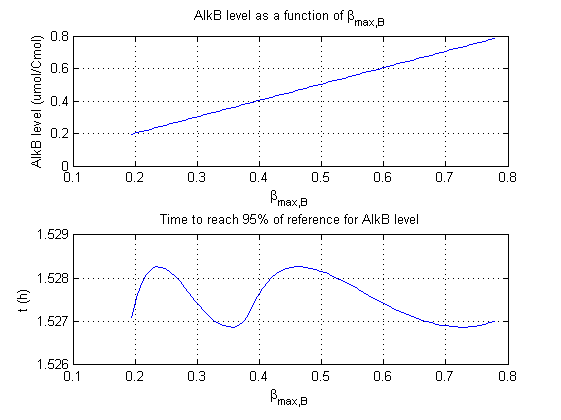
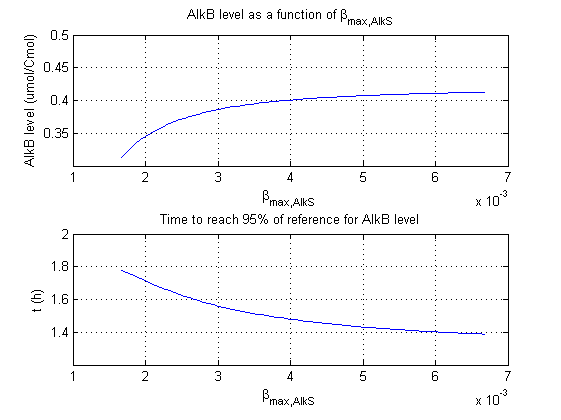
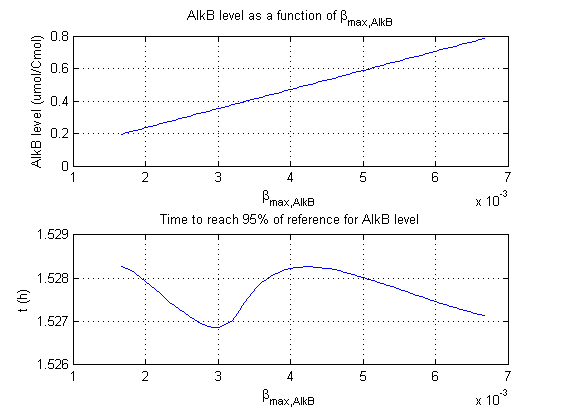
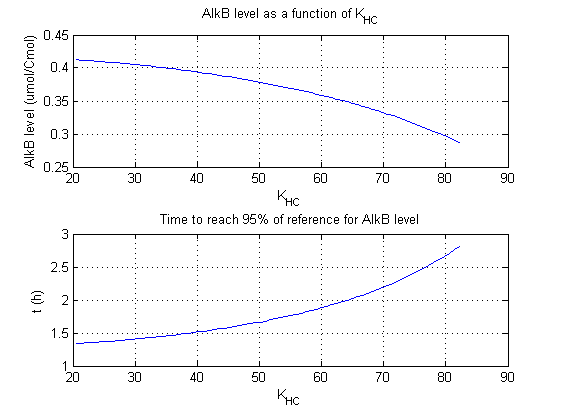
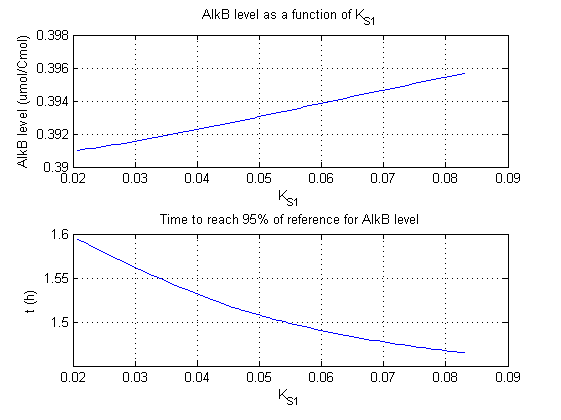
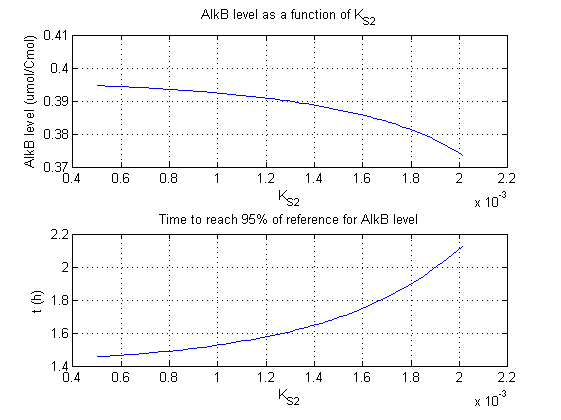
For all beta parameters (lumped variables for transcriptions speed, translations speed, promoter strength and rbs strength) and all K parameters (equilibrium constant for binding of AlkS with alkanes and coefficients for the activity of promoters) a sensitivity analysis has been performed. This was done by varying the parameters from 50% of their original values to 200% of their original values.
For this range of parameter values it is checked how much the steady state concentration of AlkB is and how long it takes for 95% of the steady state concentration of AlkB to be reached from steady state values for no alkanes to steady state values for 1 µM of alkanes.
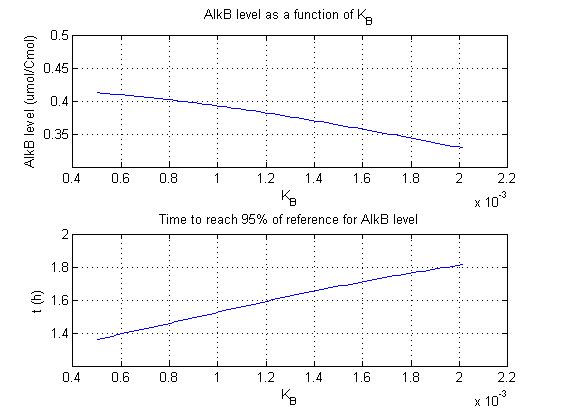
In the figures below are the results for this sensitivity analysis.
Conclusion
Most parameters have a linear or slightly exponential effect on the steady state concentration. The time it takes to reach steady state is more complexly correlated to the parameters. The wave functions for βmaxB and βmaxAlkB can be explained by the numerical integration steps. The number of time steps of the integration is finite, through interpolation the value the concentration reaches 95% of the steady state concentration is approximated. This can result however in numerical artifacts as displayed by these two functions.
These results are summarized in table 1. A '0' means insensitive, a '+' means somewhat sensitive and a '++' is very sensitive.
Table 1; results for sensitivity analysis for the gene regulation model
| Parameter | Sensitive for steady state concentration | Sensitive for reaching steady state concentration |
| βmaxS1 | 0 | + |
| βmaxS2 | + | + |
| βmaxB | ++ | 0 |
| βmaxAlkS | + | + |
| βmaxAlkB | ++ | 0 |
| KHC | + | + |
| KS1 | 0 | + |
| KS2 | + | + |
| KB | + | + |
βmaxS1 and βmaxS1 have the strongest effect on the steady state concentration of AlkB. This is logical as they directly influence the concentrations of AlkB and the mRNA of AlkB in the ODE. Interestingly they have no significant influence on how fast the system reaches steady state.
The second observation that can be made from the results is that βmaxS1 and KS1 have no significant influence on the steady state concentration of AlkB. This is logical as AlkB is expressed with higher alkane concentrations. In this situation pAlkS1 is more repressed and does not contribute much at the total AlkS and AlkB concentrations.
Go back to the model explanation here
Go back to the main modeling page here
 "
"