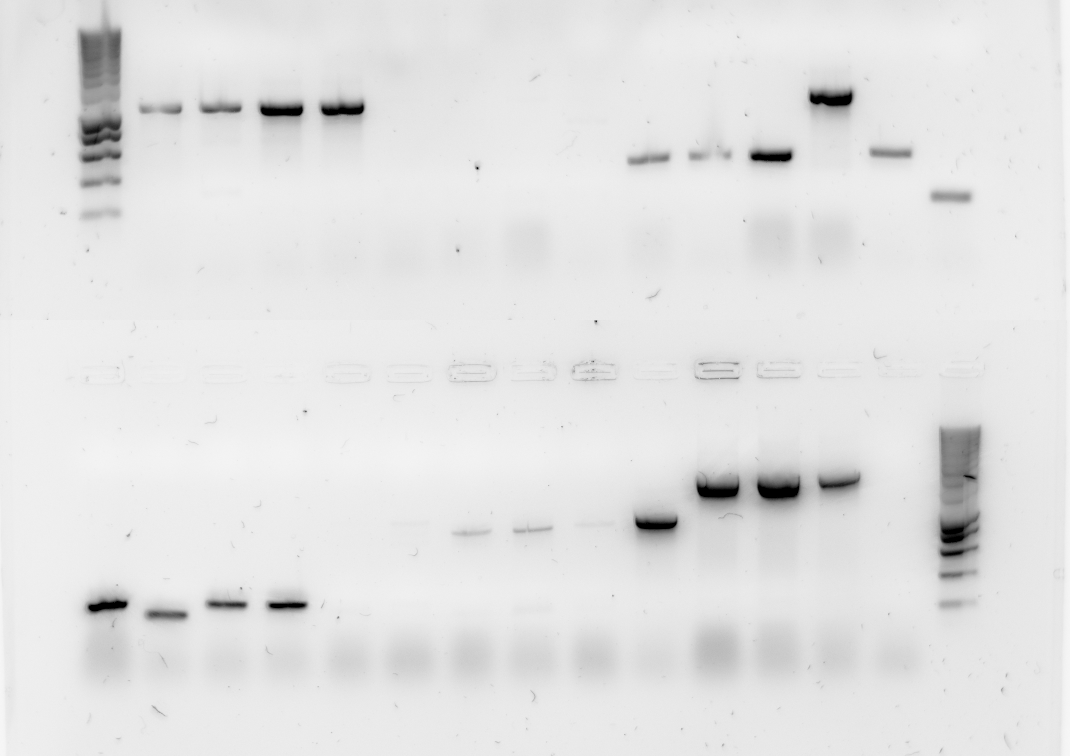Team:TU Delft/8 September 2010 content
From 2010.igem.org
Alkane degradation
Yay! Colonies on our Top10 plates! We're running a colony PCR as we speak to check if they contain the desired insert... stay tuned
So, all plates contained colonies except that of 026 (J61101-rubA4-J61107-rubR). Per plate 5 colonies were PCRed. The colony PCR showed that of 028, 108, and 331 at least one is correct. 027 didn't show any PCR product, but seeing there were more than 5 colonies on the plate we will PCR some more colonies over night - possibly there is a positive one nonetheless.
Lane description
| # | Description | Expected Length (bp) | Correct? |
| 0 | Smartladder (5μl) | ||
| 1-4 | 108 (#2-5) | 1085bp | #2,3,4,5 |
| 5-8 | 027 (#1-5) | 1644bp | None (PCR more colonies) |
| 9-13 | 028 (#1-5) | 1714bp | #4 |
| 14 | 331 (#1) | ||
| 15-18 | 331 (#2-5) | ||
| 19-23 | 306 (#1-5) | n/a | Is in another backbone (used wrong primers) |
| 24 | 108 (#1) | 1085bp | #1 |
| 25-27 | 021(A-B-C)(Hugo) | ||
| 28 | 027 (#5) | 1644 | none |
| 29 | Smartladder (5μl) |
Emulsifier
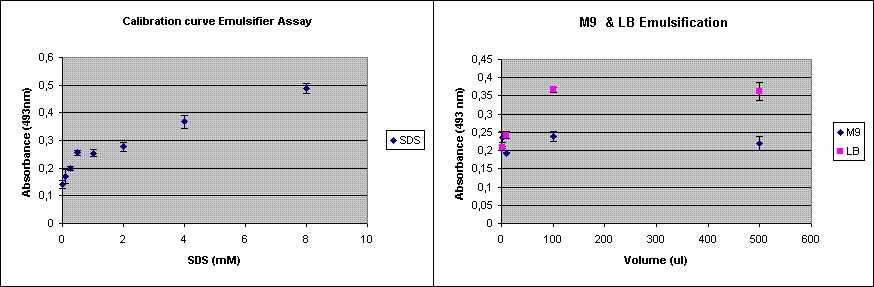
Our new construct with AlnA should be shipped by now by MrGene. Meanwhile we have optimized the protocol of the emulsification assay. In addition to the assay done last week, I now repeated the assay using only M9 and LB. This was done to see whether the medium itself already shows some background in the assay.
The experiment was done in triplicate.
Since the background was already 0.223, opposed to about 0.12 last time, the emulsification capacity of M9 seems to be in the 0.5 mM SDS range. LB shows a higher emulsification capacity. So we decided to use K12 as the host for our emulsification production.
 "
"