Team:TU Delft/18 August 2010 content
From 2010.igem.org
(Difference between revisions)
| Line 61: | Line 61: | ||
|1.398 | |1.398 | ||
|} | |} | ||
| + | |||
| + | ==Alkane degradation== | ||
| + | |||
| + | ====Transformation==== | ||
| + | Yesterday's ligation mixes were transformed into TOP10 competent cells and plated out onto CAM plates. | ||
| + | |||
| + | ====Ligation kinetics==== | ||
| + | Yesterday's ligation mixes were transformed into TOP10 competent cells and plated out onto CAM plates. | ||
| + | |||
| + | ====Digestions==== | ||
| + | |||
==Alkane Degradation parallel attempt== | ==Alkane Degradation parallel attempt== | ||
Revision as of 08:45, 31 August 2010
Contents |
Strain Characterization
By Hugo
I'm working in some growth curves, today I tested a different amount of inoculum than yesterday.
Measurements!!!
| Hour | Time | Glc | Glc+CC6 | C8 |
| 11:43 | 0h | 0.015 | 0.014 | 0.007 |
| 14:54 | 3h 11' | 0.042 | 0.052 | 0.066 |
| 16:00 | 4h 17' | 0.099 | 0.081 | 0.086 |
| 17:00 | 5h 17' | 0.196 | 0.128 | 0.142 |
| 18:00 | 6h 17' | 0.355 | 0.215 | 0.184 |
| 19:00 | 7h 17' | 0.555 | 0.341 | 0.275 |
| 9:43 | 22h | 1.473 | 1.366 | 1.195 |
| 11:43 | 24h | 1.459 | 1.360 | 1.398 |
Alkane degradation
Transformation
Yesterday's ligation mixes were transformed into TOP10 competent cells and plated out onto CAM plates.
Ligation kinetics
Yesterday's ligation mixes were transformed into TOP10 competent cells and plated out onto CAM plates.
Digestions
Alkane Degradation parallel attempt
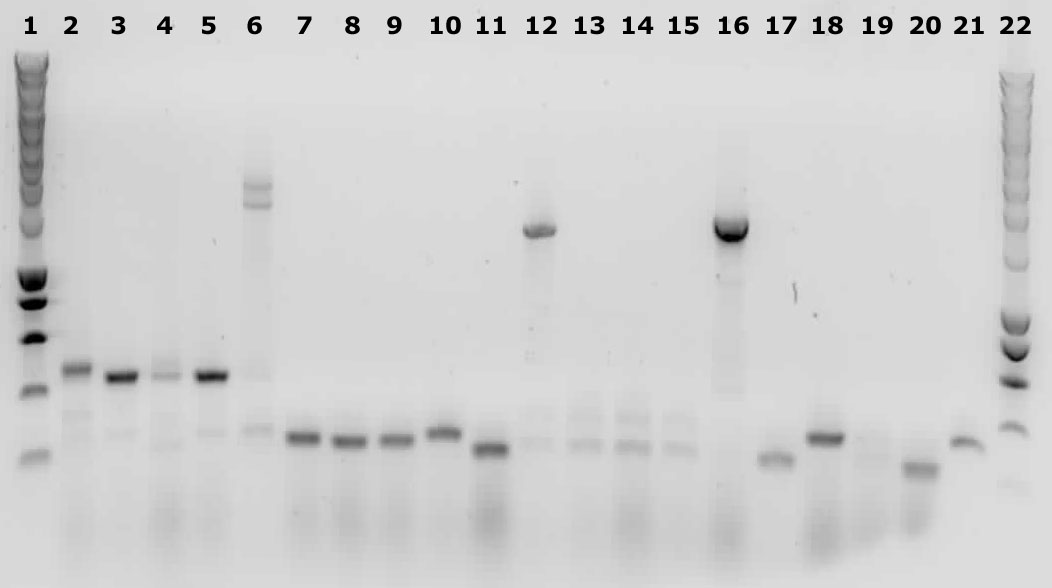
Yesterdays plates all contained colonies. But since this is a two way ligation, we expect a lot of religated backbones, so false positive colonies. That is why we performed a colony PCR.
We picked 5 colonies from each plate.
Lane description
| # | Description | Expected size (bp) | OK? |
| 1 | Smartladder | n/a | Yes |
| 2-6 | 014 | 3000+ | No |
| 7-11 | 020 | 2100+ | No |
| 12-16 | 304 | 342 | Maybe |
| 17-21 | 327 | 310 | Maybe |
Because the gels are inconclusive about biobrick 304 and 327 we decided to isolate the plasmids and send them to BaseClear for sequencing.
 "
"