Team:Stockholm/7 August 2010
From 2010.igem.org
(→Andreas) |
|||
| (5 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
* KEGG for folate: [http://www.genome.jp/kegg-bin/show_pathway?org_name=eco&mapno=00790&mapscale=&show_description=hide folate biosynthesis pathway] | * KEGG for folate: [http://www.genome.jp/kegg-bin/show_pathway?org_name=eco&mapno=00790&mapscale=&show_description=hide folate biosynthesis pathway] | ||
* Enzyme kinetics for [http://www.genome.jp/kegg-bin/show_pathway?org_name=eco&mapno=00790&mapscale=&show_description=hide folate biosynthesis pathway] better to be extracted. | * Enzyme kinetics for [http://www.genome.jp/kegg-bin/show_pathway?org_name=eco&mapno=00790&mapscale=&show_description=hide folate biosynthesis pathway] better to be extracted. | ||
| + | |||
| + | ---- | ||
| + | ==Nina== | ||
| + | ---- | ||
| + | |||
| + | ===Tranformation of site directed mutagenesis Tyrosinase=== | ||
| + | |||
| + | I transformed the site directed mutagenesis Tyrosinase in 100 ul Top 10 cells. The method was according to the procedure in protocols. However in step 1 I thawed the cells for 15 instead of 10 min. In step 2 I added 1 ul of DNA to the cells. | ||
== Andreas == | == Andreas == | ||
| Line 49: | Line 57: | ||
====Colony PCR==== | ====Colony PCR==== | ||
| - | ''PCR tubes''<br> | + | '''PCR tubes'''<br> |
*22.5 μl dH<sub>2</sub>O | *22.5 μl dH<sub>2</sub>O | ||
*1 μl VF2 | *1 μl VF2 | ||
| Line 59: | Line 67: | ||
::'' '''Negative control (18):''' Blank'' | ::'' '''Negative control (18):''' Blank'' | ||
| - | ''PCR settings'' | + | '''PCR settings''' |
{|border="1" cellpadding="2" cellspacing="0" | {|border="1" cellpadding="2" cellspacing="0" | ||
|'''1) Denaturation:''' | |'''1) Denaturation:''' | ||
| Line 89: | Line 97: | ||
'''Results'''<br> | '''Results'''<br> | ||
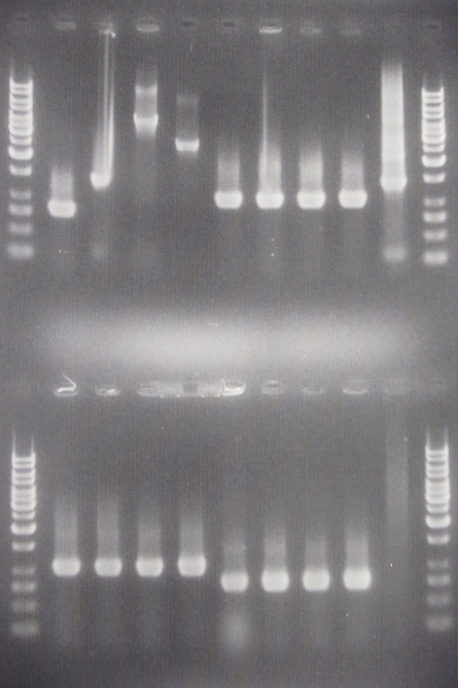
| + | [[Image:ColPCR_IgGp-SOD-yCCS_7aug.png|200px|thumb|right|Colony PCR gel verification of pSB1C3.IgG prot., pSB1A3.yCCS A & B and pSB1A3.SOD.<br> | ||
| + | '''Row 1:''' 1 kb λ, 1, 2, 3, 4, 5, 6, 7, 8, 17, 1 kb λ<br> | ||
| + | '''Row 2:''' 1 kb λ, 9, 10, 11, 12, 13, 14, 15, 16, 18, 1 kb λ<br> | ||
| + | 3 μl λ, 6 μl sample.]] | ||
Good bands for yCCS A & B and SOD, but not for IgG protease. The latter needs to be re-picked and amplified again. | Good bands for yCCS A & B and SOD, but not for IgG protease. The latter needs to be re-picked and amplified again. | ||
| Line 101: | Line 113: | ||
====Plasmid prep==== | ====Plasmid prep==== | ||
Made plasmid prep of IgG protease ON cultures set 4/8. Samples were discarded since colony PCR showed that the samples were incorrect. | Made plasmid prep of IgG protease ON cultures set 4/8. Samples were discarded since colony PCR showed that the samples were incorrect. | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 10:47, 26 October 2010
Contents |
Hassan
Vitamin B9 (folic acid)
- We are trying to find a proper model for Vitamin B9 production in E. Coli
- E. Coli lacks FolQ
- [http://www.ncbi.nlm.nih.gov/pubmed/15611104] and [http://www.ncbi.nlm.nih.gov/pubmed/17698004] could be intereseting.
- KEGG for folate: [http://www.genome.jp/kegg-bin/show_pathway?org_name=eco&mapno=00790&mapscale=&show_description=hide folate biosynthesis pathway]
- Enzyme kinetics for [http://www.genome.jp/kegg-bin/show_pathway?org_name=eco&mapno=00790&mapscale=&show_description=hide folate biosynthesis pathway] better to be extracted.
Nina
Tranformation of site directed mutagenesis Tyrosinase
I transformed the site directed mutagenesis Tyrosinase in 100 ul Top 10 cells. The method was according to the procedure in protocols. However in step 1 I thawed the cells for 15 instead of 10 min. In step 2 I added 1 ul of DNA to the cells.
Andreas
Cloning
Continued from 4/8
White colonies picked from 4/8 plates.
| Colony PCR samples | ||||
|---|---|---|---|---|
| pSB1C3.IgG prot. | A (1) | B (2) | C (3) | D (4) |
| pSB1A3.yCCS A | A (5) | B (6) | C (7) | D (8) |
| pSB1A3.yCCS B | A (9) | B (10) | C (11) | D (12) |
| pSB1A3.SOD | A (13) | B (14) | C (15) | D (16) |
Colony PCR
PCR tubes
- 22.5 μl dH2O
- 1 μl VF2
- 1 μl VR
- 0.5 μl cell suspension
- illustra Ready-to-Go PCR Beads
- Positive control (17): pSB1A3.BBa_J04450
- Negative control (18): Blank
- Positive control (17): pSB1A3.BBa_J04450
PCR settings
| 1) Denaturation: | 95 °C - 10:00 | |
| 2) Denaturation: | 95 °C - 0:30 | x30 |
| 3) Annealing: | 55 °C - 0:30 | |
| 4) Elongation: | 72 °C - 1:45 | |
| 5) Finish: | 25 °C - ∞ | |
Gel verification
1 % agarose, 70 V, 1 h
Expected bands (plus 333 bp from vector)
- IgG protease: 930 bp + 333 bp = 1263 bp
- yCCS: 744 bp + 333 bp = 1077 bp
- SOD: 459 bp + 333 bp = 792 bp
- BBa_J04450: 1069 bp + 317 bp = 1386 bp
Results
Good bands for yCCS A & B and SOD, but not for IgG protease. The latter needs to be re-picked and amplified again.
Sine very strange sequencing results from yCCS A, where the sequence was that of mCherry RFP (BBa_J18932) rather than yCCS, makes me hesitant to whether samples 5-8 and 9-12 are indeed yCCS; since yCCS and mCherry nucleotide sequences are quite similar in length.
I will run a dedicated gel tomorrow to compare yCCS with BBa_J18932 directly.
ON cultures
Clones A & B from yCCS A, yCCS B and SOD were picked for glycerol stock and plasmid prep. ON cultures were set:
- 5 ml LB + 100 Amp (37 °C, 250 rpm)
- 3 ml LB + 100 Amp (30 °C)
Plasmid prep
Made plasmid prep of IgG protease ON cultures set 4/8. Samples were discarded since colony PCR showed that the samples were incorrect.
 |

|
 |

|
 |

|

|

|
 "
"