Team:Stockholm/6 September 2010
From 2010.igem.org
(→Mimmi) |
(→Andreas) |
||
| Line 34: | Line 34: | ||
==Andreas== | ==Andreas== | ||
| + | |||
| + | ===Cloning of N-CPPs into pSB1C3=== | ||
| + | ====Transformation results==== | ||
| + | ''From 3/9'' | ||
| + | |||
| + | More growth on the 2 μl plate than on the 5 μl plate, indicating what I already suspected: contamination in transformation. Discarded both plates and restarted cloning. | ||
| + | |||
| + | ====Digestion==== | ||
| + | |||
| + | [N-CPP plasmid] = 672 ng/μl | ||
| + | |||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | | | ||
| + | !width="50"|N-CPP | ||
| + | |- | ||
| + | |10X buffer Tango | ||
| + | |align="center"|2 | ||
| + | |- | ||
| + | |DNA (1 μg) | ||
| + | |align="center"|1.5 | ||
| + | |- | ||
| + | |dH<sub>2</sub>O | ||
| + | |align="center"|12.5 | ||
| + | |- | ||
| + | |XbaI | ||
| + | |align="center"|1 | ||
| + | |- | ||
| + | |AgeI | ||
| + | |align="center"|4 | ||
| + | |- | ||
| + | | | ||
| + | !20 μl | ||
| + | |} | ||
| + | |||
| + | :Incubation: 37 °C, 3 h<br /> | ||
| + | :Inactivation: 80 °C, 20 min | ||
| + | |||
| + | ====Control digestions==== | ||
| + | Did two control digestions to analyze the N-CPP plasmid vector size. | ||
| + | |||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | | | ||
| + | !width="50"|A+S | ||
| + | !width="50"|ApaI | ||
| + | |- | ||
| + | |10X buffer Tango | ||
| + | |align="center"|2 | ||
| + | |align="center"|2 | ||
| + | |- | ||
| + | |DNA | ||
| + | |align="center"|1 | ||
| + | |align="center"|1 | ||
| + | |- | ||
| + | |dH<sub>2</sub>O | ||
| + | |align="center"|15 | ||
| + | |align="center"|16 | ||
| + | |- | ||
| + | |FD ApaI | ||
| + | |align="center"|1 | ||
| + | |align="center"|1 | ||
| + | |- | ||
| + | |FD SmaI | ||
| + | |align="center"|1 | ||
| + | |align="center"|0 | ||
| + | |- | ||
| + | | | ||
| + | !20 μl | ||
| + | !20 μl | ||
| + | |} | ||
| + | |||
| + | :Incubation: 37 °C, 30 min<br /> | ||
| + | :Inactivation: 65 °C, 5 min | ||
| + | |||
| + | =====Gel verification===== | ||
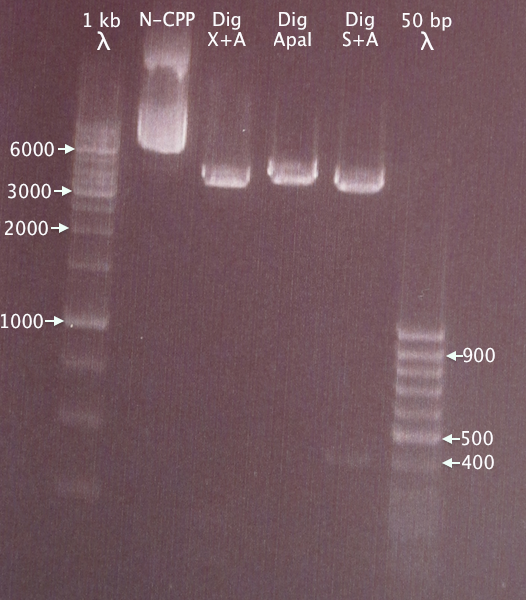
| + | [[image:Gelver NCPP dig 6sep.png|200px|right|thumb|'''Gel verification of N-CPP plasmid digestions.'''<br />1 kb λ = O'GeneRuler 1 kb DNA ladder; 50 bp λ = GeneRuler 50 bp DNA ladder.]] | ||
| + | 1 % agarose, 120 V, 40 min | ||
| + | |||
| + | *'''N-CPP:''' undigested N-CPP plasmid | ||
| + | *'''Dig X+A:''' N-CPP plasmid digested with XbaI & AgeI | ||
| + | *'''Dig ApaI:''' Linearized N-CPP plasmid digested with ApaI | ||
| + | *'''Dig S+A:''' N-CPP plasmid digested with SmaI and ApaI, excising the 391 bp N-CPP cluster. | ||
| + | |||
| + | '''Results'''<br /> | ||
| + | Linearized plasmid seems to be ≈5 kb long. Undigested plasmid moved slower; this was probably the result of the lane being too crowded. | ||
| + | |||
| + | ====Ligation==== | ||
| + | |||
| + | Vector: Dig pSB1C3 X+A EXTR 1 | ||
| + | |||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | | | ||
| + | !width="50"|1 | ||
| + | !width="50"|2* | ||
| + | |- | ||
| + | |5X Rapid Ligation buffer | ||
| + | |align="center"|4 | ||
| + | |align="center"|4 | ||
| + | |- | ||
| + | |Vector DNA | ||
| + | |align="center"|4 | ||
| + | |align="center"|4 | ||
| + | |- | ||
| + | |Insert DNA | ||
| + | |align="center"|11 | ||
| + | |align="center"|9 | ||
| + | |- | ||
| + | |dH<sub>2</sub>O | ||
| + | |align="center"|0 | ||
| + | |align="center"|2 | ||
| + | |- | ||
| + | |T4 DNA ligase | ||
| + | |align="center"|1 | ||
| + | |align="center"|1 | ||
| + | |- | ||
| + | | | ||
| + | !22 μl † | ||
| + | !20 μl | ||
| + | |} | ||
| + | † ''Accidentally added 2 μl FastAP alkaline phosphatase to ligation mix 1. Enzyme inactivated at 75 °C, 10 min, prior to ligation.'' | ||
| + | *Incubation: 22 °C, 10 min | ||
| + | |||
| + | ====Transformation==== | ||
| + | ''Performed by Mimmi'' | ||
| + | |||
| + | Standard transformation, except: | ||
| + | *30 min incubation in 37 °C | ||
| + | *3 μl ligation mix | ||
| + | Cells grown on Cm 25 plates. | ||
| + | |||
| + | ====PCR==== | ||
| + | Since we've experienced some problems with our N-CPP clonings, a PCR of each individual N-CPP was performed for individual cloning, in case the current cloning doesn't work. | ||
| + | |||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | !colspan="3"|Reactions & primers | ||
| + | |- | ||
| + | | | ||
| + | !Forward | ||
| + | !Reverse | ||
| + | |- | ||
| + | |'''N-Tra10''' | ||
| + | |align="center"|VF2 | ||
| + | |align="center"|VR | ||
| + | |- | ||
| + | |'''N-TAT''' | ||
| + | |align="center"|pEXf | ||
| + | |align="center"|pEXr | ||
| + | |- | ||
| + | |'''N-LMWP''' | ||
| + | |align="center"|pGexf | ||
| + | |align="center"|pGexr | ||
| + | |- | ||
| + | |'''N-CPP cluster''' | ||
| + | |align="center"|VF2 | ||
| + | |align="center"|pGexr | ||
| + | |} | ||
| + | <br /> | ||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | !colspan="2"|PCR tubes | ||
| + | |- | ||
| + | |10X Pfu buffer | ||
| + | |align="center" width="50"|5 | ||
| + | |- | ||
| + | |dNTP, 10 mM | ||
| + | |align="center"|1 | ||
| + | |- | ||
| + | |dH<sub>2</sub>O | ||
| + | |align="center"|41 | ||
| + | |- | ||
| + | |Template DNA | ||
| + | |align="center"|0.5 | ||
| + | |- | ||
| + | |Forw. primer | ||
| + | |align="center"|1 | ||
| + | |- | ||
| + | |Rev. primer | ||
| + | |align="center"|1 | ||
| + | |- | ||
| + | |Pfu DNA pol. | ||
| + | |align="center"|0.5 | ||
| + | |- | ||
| + | | | ||
| + | !50 μl | ||
| + | |} | ||
| + | |||
| + | Standard colony PCR settings | ||
| + | *Elongation: 1:15 | ||
| + | |||
| + | Tubes stored in -20 °C. | ||
Revision as of 13:29, 7 September 2010
Contents |
Mimmi
MITF-M
Gel
| well | sample |
|---|---|
| 1 | 1kb ladder |
| 2 | MITF-M 1 |
| 3 | MITF-M 2 |
| 4 | MITF-M 3 |
| 5 | MITF-M 4 |
| 6 | positive control |
| 7 | blank |
Andreas
Cloning of N-CPPs into pSB1C3
Transformation results
From 3/9
More growth on the 2 μl plate than on the 5 μl plate, indicating what I already suspected: contamination in transformation. Discarded both plates and restarted cloning.
Digestion
[N-CPP plasmid] = 672 ng/μl
| N-CPP | |
|---|---|
| 10X buffer Tango | 2 |
| DNA (1 μg) | 1.5 |
| dH2O | 12.5 |
| XbaI | 1 |
| AgeI | 4 |
| 20 μl |
- Incubation: 37 °C, 3 h
- Inactivation: 80 °C, 20 min
Control digestions
Did two control digestions to analyze the N-CPP plasmid vector size.
| A+S | ApaI | |
|---|---|---|
| 10X buffer Tango | 2 | 2 |
| DNA | 1 | 1 |
| dH2O | 15 | 16 |
| FD ApaI | 1 | 1 |
| FD SmaI | 1 | 0 |
| 20 μl | 20 μl |
- Incubation: 37 °C, 30 min
- Inactivation: 65 °C, 5 min
Gel verification
1 % agarose, 120 V, 40 min
- N-CPP: undigested N-CPP plasmid
- Dig X+A: N-CPP plasmid digested with XbaI & AgeI
- Dig ApaI: Linearized N-CPP plasmid digested with ApaI
- Dig S+A: N-CPP plasmid digested with SmaI and ApaI, excising the 391 bp N-CPP cluster.
Results
Linearized plasmid seems to be ≈5 kb long. Undigested plasmid moved slower; this was probably the result of the lane being too crowded.
Ligation
Vector: Dig pSB1C3 X+A EXTR 1
| 1 | 2* | |
|---|---|---|
| 5X Rapid Ligation buffer | 4 | 4 |
| Vector DNA | 4 | 4 |
| Insert DNA | 11 | 9 |
| dH2O | 0 | 2 |
| T4 DNA ligase | 1 | 1 |
| 22 μl † | 20 μl |
† Accidentally added 2 μl FastAP alkaline phosphatase to ligation mix 1. Enzyme inactivated at 75 °C, 10 min, prior to ligation.
- Incubation: 22 °C, 10 min
Transformation
Performed by Mimmi
Standard transformation, except:
- 30 min incubation in 37 °C
- 3 μl ligation mix
Cells grown on Cm 25 plates.
PCR
Since we've experienced some problems with our N-CPP clonings, a PCR of each individual N-CPP was performed for individual cloning, in case the current cloning doesn't work.
| Reactions & primers | ||
|---|---|---|
| Forward | Reverse | |
| N-Tra10 | VF2 | VR |
| N-TAT | pEXf | pEXr |
| N-LMWP | pGexf | pGexr |
| N-CPP cluster | VF2 | pGexr |
| PCR tubes | |
|---|---|
| 10X Pfu buffer | 5 |
| dNTP, 10 mM | 1 |
| dH2O | 41 |
| Template DNA | 0.5 |
| Forw. primer | 1 |
| Rev. primer | 1 |
| Pfu DNA pol. | 0.5 |
| 50 μl | |
Standard colony PCR settings
- Elongation: 1:15
Tubes stored in -20 °C.
 "
"