Team:Stockholm/5 October 2010
From 2010.igem.org
m |
|||
| Line 140: | Line 140: | ||
| his.SOD sup. | | his.SOD sup. | ||
|} | |} | ||
| + | |||
| + | ==Johan== | ||
| + | |||
| + | ===What went wrong=== | ||
| + | I got NO colonies on all plates, something is wrong. | ||
| + | ====Cut==== | ||
| + | * vector with insert, to see if its ligated | ||
| + | * vector with insert + BamHI | ||
| + | * vector with insret - BamHI | ||
| + | * 2 µl pEX vector | ||
| + | |||
| + | [[Image:SU 5oktgels.png]] | ||
| + | |||
| + | While running, I talked to Rickard, the guy that yesterday told me that it works to use invitrogen ligase enzyme with fermentas buffer. Further research showed that invitrogen buffer has Pog8000 which fermentas buffer dont have, which probably means that invitrogen ligase needs Pog8000 - which it didnt have in the fermentas buffer | ||
| + | |||
| + | ====Ligation & transformation==== | ||
| + | Did another ligation into pEX with fermentas ligase AND buffer, I then transformed 3 µl of all ligations into top10 cells. | ||
| + | |||
| + | ====Coomassie==== | ||
| + | Did a coomassie of some constructs from Andreas & Mimmi. SOD, tat-SOD-his, SOD-his, his-SOD and yCCS. 0, 1, 2 and 3h after IPTG induction for all samples. | ||
{{Stockholm/Footer}} | {{Stockholm/Footer}} | ||
Latest revision as of 20:47, 27 October 2010
Contents |
Andreas
Transfer of pEX.nCPP⋅SOD⋅His to BL21
Glycerol stocks
- BL21 pEX.nLMWP⋅SOD⋅His
- BL21 pEX.nTra10⋅SOD⋅His
Assembly of His⋅SOD⋅cCPP constructs
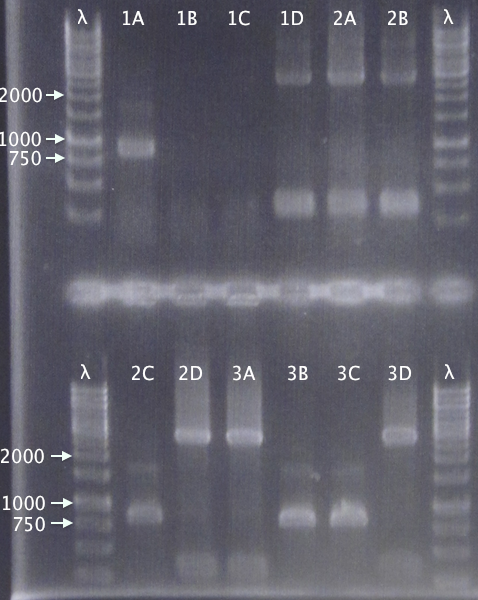
Colony PCR
- pSB1C3.His⋅SOD⋅cTra10 A-D
- pSB1C3.His⋅SOD⋅cTAT A-D
- pSB1C3.His⋅SOD⋅cLMWP A-D
Standard colony PCR settings
- Elongation time: 1:30
Gel verification
1 % agarose, 120 V
Expected bands
- 884 bp
- 854 bp
- 863 bp
Results
Varying band sized, with the following clones seeming correct:
- A
- D
- B & C
Transfer of nCPP⋅SOD⋅His.RBS.yCCS operon to pEX
Plasmid preps
From 4/10 ON cultures
As I'm still waiting for sequencing results of the operons in pSB1K3, I spun down cells (4,400 x g, 10 min), and saved pellets in -20 °C ON.
- pEX.nTAT⋅SOD⋅His.RBS.yCCS 2: A
- pEX.nTAT⋅SOD⋅His.RBS.yCCS 3: B
- pEX.nTra10⋅SOD⋅His.RBS.yCCS 1: B
- pEX.nTra10⋅SOD⋅His.RBS.yCCS 2: B
- pEX.nLMWP⋅SOD⋅His.RBS.yCCS 2: B
- pEX.nLMWP⋅SOD⋅His.RBS.yCCS 3: B
Nina
Ligation of proteins into peX and CPP containing vectors
Ligations:
- Fusion protein-His into three vectors each containing all N-terminal CPPs LMWP, TAT and Tra10.
* Gene: 10 ul * Vector: LMWP 0.5 ul (25ng), TAT 1 ul (20 ng) and Tra10 1 ul (20 ng) * Quick Ligation buffer 2X: LMWP 11.5 ul, TAT 12 ul and Tra10 12 ul * Quick Ligase: 1 ul
- His-Fusion protein into three vectors each containing all C-terminal CPPs LMWP, TAT and Tra10.
* Gene: 6 ul * Vector: CPP1 0.5 ul (25ng), TAT 1 ul (20 ng) and CPP3 1 ul (20 ng) * Quick Ligation buffer 2X: CPP1 7.5 ul, TAT 8 ul and CPP3 8 ul * Quick Ligase: 1 ul
- Protein A-His into three vectors each containing all N-terminal CPPs LMWP, TAT and Tra10.
* Gene: 12.5 ul * Vector: LMWP 0.5 ul (25ng), TAT 1 ul (20 ng) and Tra10 1 ul (20 ng) * Quick Ligation buffer 2X: LMWP 14 ul, TAT 14.5 ul and Tra10 14.5 ul * Quick Ligase: 1 ul
- IgG protease_Tra10_Ntermin_#6 into peX vector
* Gene: 7 ul * Vector: peX 1 (25ng) * Quick Ligation buffer 2X: 9 ul * Quick Ligase: 1 ul
All ligation mixtures were incubated in 22 °C (in a water bath) for 15 minutes. During these minutes I also thawed competent Top 10 cells (100 ul) on ice for transformation of the ligation samples.
Transformation of ligation products
The procedure was according to the method decribed in protocols. However I thawed Top 10 cells (100 ul) in 15 minutes instead of 10. I added 3 ul of ligation samples into each 100 of Top 10 cells.
Mimmi
Protein purification
- Resuspend pellet in
| mix | ||
|---|---|---|
| 626.85µl | NPI-10/20 | |
| 3.15µl | imidazole 2M |
- Add 70µl lysozyme
- Add 14µl DNase 20µg/ml -> 14µg
- Add 0.7µl PMSF 1mM
- Inbcubate on ice for 15-30min
- Centrifuge lysate at 12,000xg for 20min at 4°C
- Collect supernatant
- Save 20µl for SDS-Page
- PhastGel
| well | sample | |
|---|---|---|
| 1 | ladder | |
| 2 | SOD 3h | |
| 3 | SOD.his 0h | |
| 4 | SOD.his sup. | |
| 5 | his.SOD 0h | |
| 6 | his.SOD sup. |
Johan
What went wrong
I got NO colonies on all plates, something is wrong.
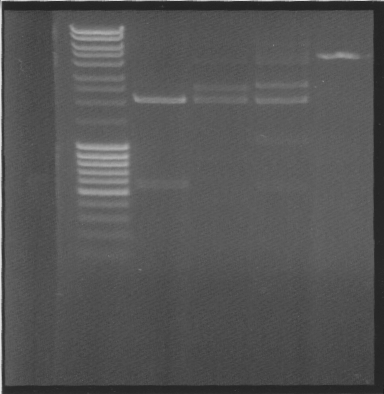
Cut
- vector with insert, to see if its ligated
- vector with insert + BamHI
- vector with insret - BamHI
- 2 µl pEX vector
While running, I talked to Rickard, the guy that yesterday told me that it works to use invitrogen ligase enzyme with fermentas buffer. Further research showed that invitrogen buffer has Pog8000 which fermentas buffer dont have, which probably means that invitrogen ligase needs Pog8000 - which it didnt have in the fermentas buffer
Ligation & transformation
Did another ligation into pEX with fermentas ligase AND buffer, I then transformed 3 µl of all ligations into top10 cells.
Coomassie
Did a coomassie of some constructs from Andreas & Mimmi. SOD, tat-SOD-his, SOD-his, his-SOD and yCCS. 0, 1, 2 and 3h after IPTG induction for all samples.
 |

|
 |

|
 |

|

|

|
 "
"