Team:Stockholm/4 October 2010
From 2010.igem.org
m |
|||
| Line 284: | Line 284: | ||
| 10ml | | 10ml | ||
|} | |} | ||
| + | |||
| + | ==Johan== | ||
| + | |||
| + | ===Miniprep=== | ||
| + | |||
| + | tra10-bFGF-his, tat-bFGF-his, tat-bFGF-his, lmwp-bFGF-his, his-bFGF-tra10, his-bFGF-lmwp | ||
| + | |||
| + | All ~300 ng/µl | ||
| + | |||
| + | ===Digest miniprep=== | ||
| + | |||
| + | 2 µl DNA | ||
| + | |||
| + | (1 µl BamHI) | ||
| + | |||
| + | 2 µl 10x buffer | ||
| + | |||
| + | 15 µl H2O | ||
| + | |||
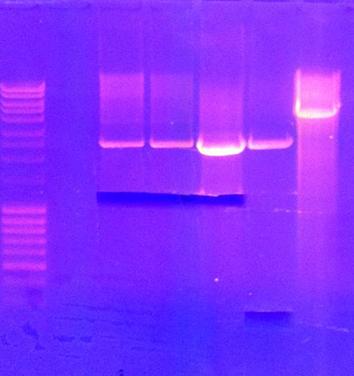
| + | ===Gel=== | ||
| + | |||
| + | tra10-bFGF-his, tat-bFGF-his, tat-bFGF-his, lmwp-bFGF-his, his-bFGF-tra10, his-bFGF-lmwp | ||
| + | |||
| + | [[Image:SU 4oktgels.png]] | ||
| + | |||
| + | Results: All constructs had one correct miniprep | ||
| + | |||
| + | ===Cut=== | ||
| + | |||
| + | The parts was cut from its vector to be ligated into pEX vector | ||
| + | |||
| + | 5 µl bFGF | ||
| + | |||
| + | 2 µl 10x buffer | ||
| + | |||
| + | 1 µl XbaI | ||
| + | |||
| + | 1 µl PstI | ||
| + | |||
| + | 11 µl H2O | ||
| + | |||
| + | ===Ligation=== | ||
| + | |||
| + | The vector was first treated with alkaline phosphatase for 60 min. | ||
| + | |||
| + | 7 µl bFGF | ||
| + | |||
| + | 1 µl pEX | ||
| + | |||
| + | 2 µl 10x buffer | ||
| + | |||
| + | 1 µl ligase | ||
| + | |||
| + | 9 µl H2O | ||
| + | |||
| + | ===Transformation=== | ||
| + | |||
| + | 3 µl from all constructs was then transformed into top10 cells. | ||
| + | |||
| + | ===Coomassie gels=== | ||
| + | |||
| + | I made 2 coomassie gels to be used tomorrow | ||
{{Stockholm/Footer}} | {{Stockholm/Footer}} | ||
Latest revision as of 21:05, 27 October 2010
Contents |
Andreas
Transfer of pEX.nCPP⋅SOD⋅His to BL21
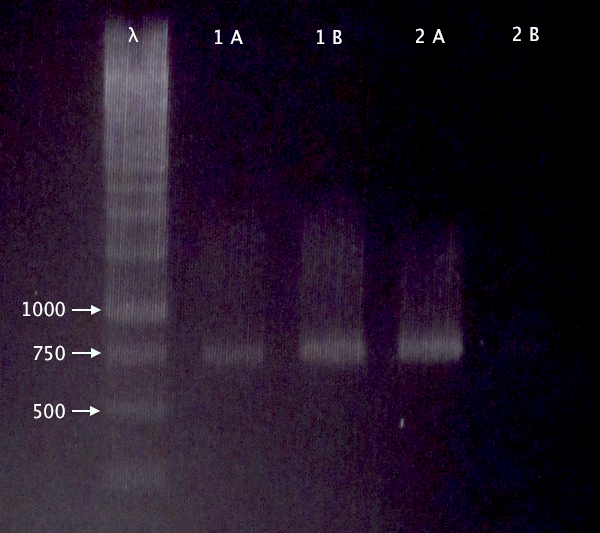
Gel verification
Re-run of 2/10 BL21 samples
- BL21 pEX.nLMWP⋅SOD⋅His: A & B
- BL21 pEX.nTra10⋅SOD⋅His: A & B
1 % agarose, 120 V
Expected bands:
- 744 bp
- 765 bp
Results
- Both clones verified
- Clone A verified; weak band for clone B
ON cultures
- 3 ml LB, 30 °C
- A (BL21 pEX.nLMWP⋅SOD⋅His)
- A (BL21 pEX.nTra10⋅SOD⋅His)
Transfer of nCPP⋅SOD⋅His.RBS.yCCS operon to pEX
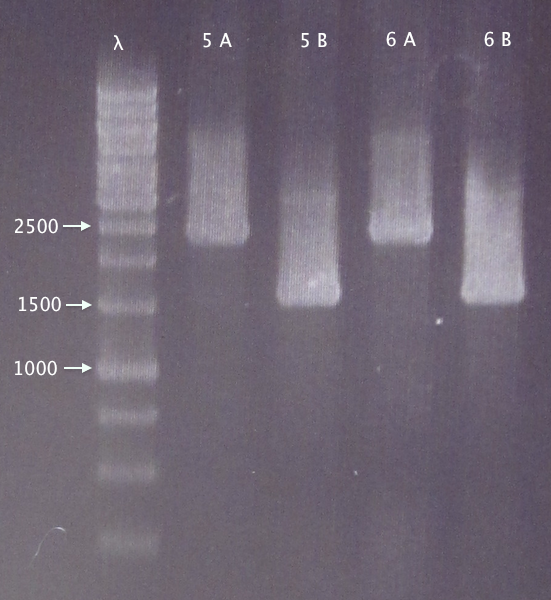
Colony PCR
Picked 2 new colonies of each of the two constructs transformed 30/8:
- 5. pEX.nTra10⋅SOD⋅His.RBS.yCCS 1: A & B
- 6. pEX.nTra10⋅SOD⋅His.RBS.yCCS 2: A & B
Standard colony PCR settings
- Elongation time: 2:00
Gel verification
0.8 % agarose, 100 V
Expected bands:
- 5. 1553 bp
- 6. 1553 bp
Results
- 5. Relevant band for clone B; too large insert (double?) for clone A.
- 6. Relevant band for clone B; too large insert (double?) for clone A.
ON cultures
- 5 ml LB, 37 °C, 250 rpm
- pEX.nTAT⋅SOD⋅His.RBS.yCCS 2: A
- pEX.nTAT⋅SOD⋅His.RBS.yCCS 3: B
- pEX.nTra10⋅SOD⋅His.RBS.yCCS 1: B
- pEX.nTra10⋅SOD⋅His.RBS.yCCS 2: B
- pEX.nLMWP⋅SOD⋅His.RBS.yCCS 2: B
- pEX.nLMWP⋅SOD⋅His.RBS.yCCS 3: B
Verification of pSB1x3 plasmids
Due to some strange growth results with our stock plasmids (pSB1x3.BBa_J04450), I decided to verify their antibiotic resistance. Restreaked clones of the following plasmids (w/ BBa_J04450 inserts) onto Amp 100, Km 50 and Cm 25 plates:
- pSB1A3
- pSB1C3
- pSB1K3
- pSB1AC3
- pSB1AK3
Assembly of His⋅SOD⋅cCPP constructs
Continued from 1/10
Received cCPPs (cTra10, cTAT and cLMWP) in pSB1C3 plasmids, digested with EcoRI and NgoMIV, from Johan.
Ligations
- Vectors:
- Dig pSB1C3.cTra10 E+N
- Dig pSB1C3.cTAT E+N
- Dig pSB1C3.cLMWP E+N
- Insert: Dig pMA.His⋅SOD E+A
| 1 | 2 | 3 | |
|---|---|---|---|
| 10X T4 Ligase buffer | 2 | 2 | 2 |
| Vector DNA | 1 | 1 | 1 |
| Insert DNA | 5 | 5 | 5 |
| dH2O | 11 | 11 | 11 |
| T4 DNA ligase | 1 | 1 | 1 |
| 20 μl | 20 μl | 20 μl |
- Incubation: 22 °C, 15 min
Transformations
- pSB1C3.His⋅SOD⋅cTra10
- pSB1C3.His⋅SOD⋅cTAT
- pSB1C3.His⋅SOD⋅cLMWP
- Standard transformation
- 1 μl
- Cm 25
Nina
Mini prep on IgG_Tra10_N#6
I performed a mini prep on yesterday's inoculated IgG_Tra10_N colony # 6 in 12 ml LB with 24 ul chloramphenicol. The procedure was according to the method described in protocols.
Overday culture of SOD
I put an overday culture of SOD-peX, His-SOD, SOD-His, SOD_TAT_Ntermin and as a positive control yCC-peX from Andrea's glycerol stocks in 12 ml LB with 24 ul amphicillin. I inoculated with a big pipett tip amount in order to have the culture reaching OD 0.6 during the day of laboration.
The reason for why I did this culture was because I had talked to Mimmi about her overexpressions about these constructs and she hadn't got any satisfying results on her gels beside from the yCC sample (which became my positive control). I wanted therefore to check if I would also obtain her type of outcome when overexpressing the proteins of interest.
Gene digestions
I performed digestions of my genes of interest in order to do a gel clean up and follow with a ligation that should yield many positive cloning results.
Digestions:
- Fusion_EA_His # 1 & 3 10 ul
- Fast digestion buffer 10 X 2 ul
- H2O 6 ul
- Restriction enzyme NgoMVI 1 ul
- Restriction enzyme PstI 1 ul (Added after 1.5 h in 37 °C)
- Fusion_NS_His # 2 10 ul
- Fast digestion buffer 10 X 2 ul
- H2O 6 ul
- Restriction enzyme AgeI 1 ul
- Restriction enzyme EcoRI 1 ul (Added after 1.5 h in 37 °C)
- Protein A_EA_His # 5 10 ul
- Fast digestion buffer 10 X 2 ul
- H2O 6 ul
- Restriction enzyme NgoMVI 1 ul
- Restriction enzyme PstI 1 ul (Added after 1.5 h in 37 °C)
- IgG protease_EA_His # 5 10 ul
- Fast digestion buffer 10 X 2 ul
- H2O 6 ul
- Restriction enzyme NgoMVI 1 ul
- Restriction enzyme PstI 1 ul (Added after 1.5 h in 37 °C)
- IgG protease_Tra10_Ntermin # 4 & 6 10 ul
- Fast digestion buffer 10 X 2 ul
- H2O 6 ul
- Restriction enzyme XbaI 1 ul
- Restriction enzyme PstI 1 ul
Incubate in 37 °C for 30 minutes.
Agarose gel on digests
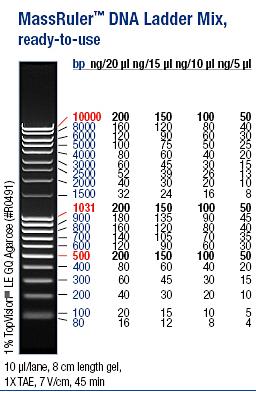
I ran an agarose gel 1 % 100 V on the digested samples in order to check if they have been digested by the enzymes and followed by cutting the bands of interest out of the gel with a scalpel over a UV-lamp for a gel clean up.
Ladder: MassRuler™ DNA Ladder Mix, ready-to-use, 80-10,000 bp
Arrangement on gels:
Gel clean up
I performed a gel clean up of the digested genes. All except IgGEA#5 were digested and cut out of the gel.
I performed the clean up according to the method described in protocols.
The measurements of the cut gel bands, addition of kit solutions and incubation time:
- All bands had a weight of aproximately 0.12 g (120 mg). 120 mg * 3 = 360 ul QXI
- I added 10 ul of QIAEXII to all samples
- All samples were incubated for 5 minutes at RT
- I performed step 11 in the procedure description
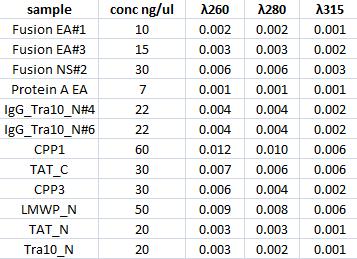
Concentration measurments
I measured with a spectrophotometer the concentration of the gel clean up samples.
Sending for sequencing
I sent IgG_Tra10_Ntermin#6 for sequencing. I mixed 15 ul of the miniprep sample and 1.5 ul of Forward bank vector verification primer (VF).
- ASB0045 898
Mimmi
Protein purification
- Using Qiagen Ni-NTA Spin Kit
Buffers
| Lysis buffer NPI-10
Wash buffer NPI-20 | NaH2PO4•H2O
NaCl + imidazole | 3.45g
8.77g 6.8g | 50mM
300mM 2M | \
/ 500ml 100ml |
|
Elution buffer | NaH2PO4•H2O
NaCl imidazole | 3.45g
8.77g 17g | 50mM
300mM 500mM | )
> 500ml ) |
| Lysosyme | 0.1g | 10mg/ml | 10ml |
Johan
Miniprep
tra10-bFGF-his, tat-bFGF-his, tat-bFGF-his, lmwp-bFGF-his, his-bFGF-tra10, his-bFGF-lmwp
All ~300 ng/µl
Digest miniprep
2 µl DNA
(1 µl BamHI)
2 µl 10x buffer
15 µl H2O
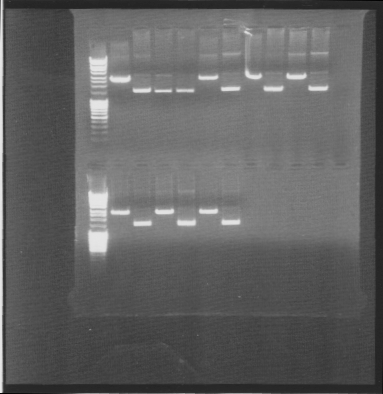
Gel
tra10-bFGF-his, tat-bFGF-his, tat-bFGF-his, lmwp-bFGF-his, his-bFGF-tra10, his-bFGF-lmwp
Results: All constructs had one correct miniprep
Cut
The parts was cut from its vector to be ligated into pEX vector
5 µl bFGF
2 µl 10x buffer
1 µl XbaI
1 µl PstI
11 µl H2O
Ligation
The vector was first treated with alkaline phosphatase for 60 min.
7 µl bFGF
1 µl pEX
2 µl 10x buffer
1 µl ligase
9 µl H2O
Transformation
3 µl from all constructs was then transformed into top10 cells.
Coomassie gels
I made 2 coomassie gels to be used tomorrow
 |

|
 |

|
 |

|

|

|
 "
"