Team:Stockholm/30 September 2010
From 2010.igem.org
m |
m |
||
| Line 341: | Line 341: | ||
**his.SOD | **his.SOD | ||
**nTAT.SOD.his | **nTAT.SOD.his | ||
| + | |||
| + | ==Johan== | ||
| + | |||
| + | ===Colony PCR screen=== | ||
| + | |||
| + | Of all constructs in the CPP-vector. tra10-bFGF-his, tat-bFGF-his, lmwp-bFGF-his, his-bFGF-tra10, his-bFGF-tat, his-bFGF-lmwp | ||
| + | |||
| + | 0,5 µl pol | ||
| + | |||
| + | 0,5 µl dNTP | ||
| + | |||
| + | 5 µl 5x buffer | ||
| + | |||
| + | 2 µl VF2 primer | ||
| + | |||
| + | 2 µl VR primer | ||
| + | |||
| + | 15 µl H2O | ||
| + | |||
| + | 40 colonies in total, 40x mastermix | ||
| + | |||
| + | ===Gel=== | ||
| + | |||
| + | [[Image:SU 30sepgels.png]] | ||
| + | |||
| + | [[Image:SU 30sepgels 1.png]] | ||
| + | |||
| + | [[Image:SU 30sepgels 2.png]] | ||
| + | |||
| + | Compilation of all constructs. All construct had at least one band of correct size. One of each and two for tat-bFGF-his was put for overnight culture. | ||
| + | |||
| + | ===Ligation for Nina=== | ||
| + | |||
| + | Did a ligation for Nina | ||
| + | |||
| + | One cut construct of IgG, and two cut constructs of Protein A. All had a concentration of ~50 ng/µl. | ||
| + | |||
| + | 19 µl insert | ||
| + | |||
| + | 0,5 µl pMA (vector with histag) -> 25 ng | ||
| + | |||
| + | 2 µl ligase | ||
| + | |||
| + | 2 µl buffer | ||
{{Stockholm/Footer}} | {{Stockholm/Footer}} | ||
Latest revision as of 21:20, 27 October 2010
Contents |
Andreas
Transfer of nCPP⋅SOD⋅His.RBS.yCCS operon to pEX
Digestions
- pSB1K3.nTAT⋅SOD⋅His.RBS.yCCS
- Clones 2 & 3
- pSB1K3.nTra10⋅SOD⋅His.RBS.yCCS
- Clones 1 & 2
- pSB1K3.nLMWP⋅SOD⋅His.RBS.yCCS
- Clones 2 & 3
| 1:2 | 1:3 | 2:1 | 2:2 | 3:2 | 3:3 | |
| 10X FastDigest buffer | 2 | 2 | 2 | 2 | 2 | 2 |
| DNA (1 μg) | 5.2 | 4.1 | 2.5 | 3.3 | 4 | 4.1 |
| dH2O | 10.8 | 11.9 | 13.5 | 12.7 | 12 | 11.9 |
| FD XbaI | 1 | 1 | 1 | 1 | 1 | 1 |
| FD PstI | 1 | 1 | 1 | 1 | 1 | 1 |
| 20 μl | 20 μl | 20 μl | 20 μl | 20 μl | 20 μl |
|---|
- Incubation: 37 °C, 1.45
- Inactivation: 80 °C, 20 min
Ligations
- Vector: [Dig pEX.RFP X+P
| pEX.1:2 | pEX.1:3 | pEX.2:1 | pEX.2:2 | pEX.3:2 | pEX.3:3 | |
| 10X T4 Ligase buffer | 2 | 2 | 2 | 2 | 2 | 2 |
| Vector DNA | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 | 1.5 |
| Insert DNA | 8 | 8 | 8 | 8 | 8 | 8 |
| dH2O | 7.5 | 7.5 | 7.5 | 7.5 | 7.5 | 7.5 |
| T4 DNA ligase | 1 | 1 | 1 | 1 | 1 | 1 |
| 20 μl | 20 μl | 20 μl | 20 μl | 20 μl | 20 μl |
|---|
- Incubation: 22 °C, 15 min
Transformation
Quick transformation
- 1 μl ligation mix
- 50 μl 0.1 M IPTG
- pEX.1:2 (pEX.nTAT⋅SOD⋅His.RBS.yCCS 2)
- pEX.1:3 (pEX.nTAT⋅SOD⋅His.RBS.yCCS 2)
- pEX.2:1 (pEX.nTra10⋅SOD⋅His.RBS.yCCS 1)
- pEX.2:2 (pEX.nTra10⋅SOD⋅His.RBS.yCCS 2)
- pEX.3:2 (pEX.nLMWP⋅SOD⋅His.RBS.yCCS 2)
- pEX.3:3 (pEX.nLMWP⋅SOD⋅His.RBS.yCCS 3)
Transformation of BL21
Quick transformation
- 50 μl competent cells
- 0.5 μl plasmid
- pEX.nTra10⋅SOD⋅His
- pEX.nLMWP⋅SOD⋅His
ON cultures
- 3 ml LB + appropriate antibiotic; 30 °C
- pEX.nTAT⋅SOD⋅His (Top10; Amp 100)
- pSB1K3.BBa_J04450 (Top10; Km 50)
Nina
Send for sequencing
I sent samples for sequencing and the mixtures were 15 ul sample and 1.5 ul forward bank vector verification primer.
- Protein A in LMWP_Ntermin ASB0045 680
- Protein A in TAT_Ntermin ASB0045 679
- Protein A in Tra10_Ntermin ASB0045 678
Digestion of protein A and peX vector
I got a mini prep of the peX vector from Andreas with a concentration of 55.52 ng/ul. I cut this vector and protein A in CPPs_N vectors.
peX:
- Fast digest buffer 10X 3.4 ul
- DNA 30 ul
- Restriction enzyme XbaI 2 ul
- Restriction enzyme PstI 2 ul
Incubated in 37 °C for 30 min.
Protein A:
- Fast digest buffer 10X 2.2 ul
- DNA 20 ul
- Restriction enzyme XbaI 1 ul
- Restriction enzyme PstI 1 ul
Incubated in 37 °C for 30 min.
Agarose gel on digests
I ran the digested products on an agarose gel 1 % 100 V.
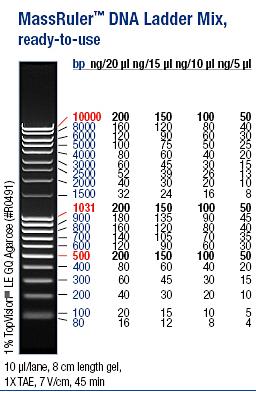
Ladder: MassRuler™ DNA Ladder Mix, ready-to-use, 80-10,000 bp
Arrangement on gel:
Gel clean up
I performed a gel clean up according to the procedures described in protocols.
On the gel it looked like it was only the peX vector that had been cut correctly and thus the protein A didn't seem to have been previously inserted correctly into the vectors holding the CPPs on the N-terminal. Therefore I only cut out and gel clean the peX vector for later use in overexpressions of our proteins.
The measurements of the cut gel bands, addition of kit solutions and incubation time:
- All bands had a weight of approximately 300 mg. 300 mg * 3 = 900 ul QXI
- I added 30 ul of QIAEXII to all samples
- All samples were incubated for 5 minutes at 50 °C
Miniprep
I performed a mini prep on Fusion EA # 1 and 3. Fusion AS I have to put a new overnight culture of since I accidentally dropped the solution and the material is now lost.
The procedure was according to the method described in protocols.
Overnight culture
I inoculated IgG protease_Tra10 # 4 & 6 in each 12 ml LB falcon tubes with 24 ul chloramphenicol.
Digestion of vectors with CPP
I digested bank vectors holding LMWP_N, TAT_N, Tra10_N, CPP1_C, TAT_C and CPP3_C to become followed by a gel clean up.
Digestion:
- DNA 20 ul
- Fast digest buffer 10X 2.2 ul
CPPs-Ntermin:
- Restriction enzyme AgeI 1 ul
- Restriction enzyme PstI 1 ul (Added after 1.5 h in 37 °C)
CPPs-Ctermin:
- Restriction enzyme NgoMIV 1 ul
- Restriction enzyme EcoRI 1 ul (Added after 1.5 h in 37 °C)
Agarose gel on digests
I ran the digests on an agarose 1 % gel 80 V.
Ladder: MassRuler™ DNA Ladder Mix, ready-to-use, 80-10,000 bp
Arragement on gel:
Gel clean up
I performed a gel clean up according to the procedure described in protocols.
The measurements of the cut gel bands, addition of kit solutions and incubation time:
- All bands had a weight of approximately 150 mg. 150 mg * 3 = 450 ul QXI
- I added 30 ul of QIAEXII to all samples
- All samples were incubated for 5 minutes at RT
Concentration measurments
I measured with a spectrophotometer the concentration of the gel clean up samples.
Mimmi
Expression
- yCCS expression looks very fine!
- no SOD
- much to low concentration loaded?
- try E-colves for more air
- taking samples after both 0.5h and 3h if the protein becomes degraded after time
SOD operon
Plasmid prep. and sequencing
- Follow E.T.Z.N.A plasmid prep. protocol
- wash 1x wash buffer
- eluate in 50µl
- Measure DNA concentration
pSB1K3.nLMWP.SOD.his.RBS.yCCS 2
pSB1K3.nLMWP.SOD.his.RBS.yCCS 3
pSB1K3.nTAT.SOD.his.RBS.yCCS 2
pSB1K3.nTAT.SOD.his.RBS.yCCS 3
pSB1K3.nTra10.SOD.his.RBS.yCCS 1
pSB1K3.nTra10.SOD.his.RBS.yCCS 2
- Send for sequencing...
Over expression
- Start ON culture
- SOD
- SOD.his
- his.SOD
- nTAT.SOD.his
Johan
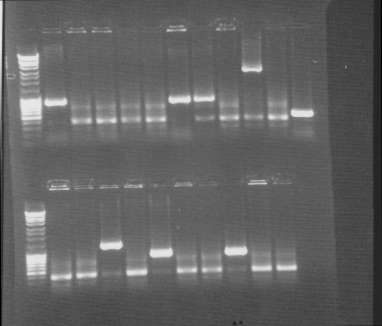
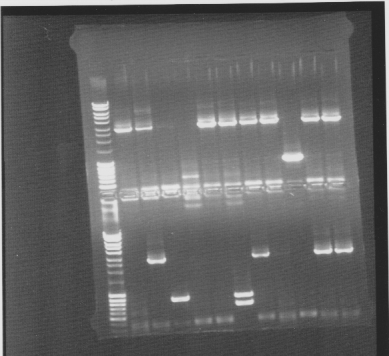
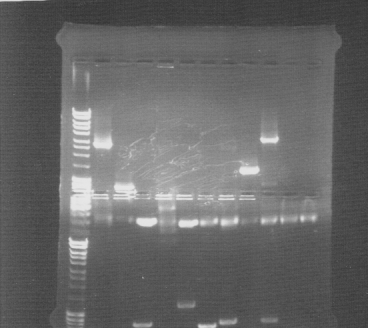
Colony PCR screen
Of all constructs in the CPP-vector. tra10-bFGF-his, tat-bFGF-his, lmwp-bFGF-his, his-bFGF-tra10, his-bFGF-tat, his-bFGF-lmwp
0,5 µl pol
0,5 µl dNTP
5 µl 5x buffer
2 µl VF2 primer
2 µl VR primer
15 µl H2O
40 colonies in total, 40x mastermix
Gel
Compilation of all constructs. All construct had at least one band of correct size. One of each and two for tat-bFGF-his was put for overnight culture.
Ligation for Nina
Did a ligation for Nina
One cut construct of IgG, and two cut constructs of Protein A. All had a concentration of ~50 ng/µl.
19 µl insert
0,5 µl pMA (vector with histag) -> 25 ng
2 µl ligase
2 µl buffer
 |

|
 |

|
 |

|

|

|
 "
"