Team:Stockholm/29 September 2010
From 2010.igem.org
(New page: {{Stockholm/Top2}} ==Andreas==) |
m |
||
| (9 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| + | |||
==Andreas== | ==Andreas== | ||
| + | |||
| + | ===Sequencing results=== | ||
| + | Sequencing results from 27/9 samples arrived and were analyzed by nucleotide BLAST. | ||
| + | |||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | !Construct | ||
| + | !Sequencing<br />result | ||
| + | !Blastn<br />result | ||
| + | !Comment | ||
| + | |- | ||
| + | |pEX.N-TAT⋅SOD⋅His 3 | ||
| + | |[[media:PEX.nTAT.SOD.his3_premix_fasta_29sep.txt|pEX.nTAT.SOD.his 3_premix]] | ||
| + | |[[media:Blastn_pEX.nTAT.SOD.his_3_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:green">Verified'''</span>. Three silent mutations in His tag; does not alter a.a. sequence. | ||
| + | |- | ||
| + | |pEX.N-TAT⋅SOD⋅His 4 | ||
| + | |[[media:PEX.nTAT.SOD.his4_premix_fasta_29sep.txt|pEX.nTAT.SOD.his 4_premix]] | ||
| + | |[[media:Blastn_pEX.nTAT.SOD.his_4_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:red;">Not verified</span>'''. Incorrect construct. | ||
| + | |- | ||
| + | |pSB1A2.RBS.yCCS 3 | ||
| + | |[[media:PSB1A2.RBS.yCCS3_premix_fasta_29sep.txt|pSB1A2.RBS.yCCS 3_premix]] | ||
| + | |[[media:Blastn_pSB1A2.RBS.yCCS3_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:orange">Partly verified</span>'''. Three mutations found: (1) Single base deletion in yCCS sequence, causing a frame shift and an extended open reading frame; (2) Insertion between SpeI and PstI, not affecting expression of cloning; (3) Point mutation in PstI site, disabling PstI restriction. Mutations (1) and (3) are both quite unsure, as the sequencing quality in this area is very low. New sequencing from VR will be sent. | ||
| + | |- | ||
| + | |pSB1A2.RBS.yCCS 4 | ||
| + | |[[media:PSB1A2.RBS.yCCS4_premix_fasta_29sep.txt|pSB1A2.RBS.yCCS 4_premix]] | ||
| + | |[[media:Blastn_pSB1A2.RBS.yCCS4_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:orange">Partly verified</span>'''. Four mutations found: (1) Point mutation changing a K codon to T; (2) Single base deletion in yCCS sequence, causing frame shift; (3) Insertion between SpeI and PstI, not affecting expression of cloning; (4) Point mutation in PstI site, disabling PstI restriction. See comment | ||
| + | |- | ||
| + | |pSB1K3.N-TAT⋅SOD⋅His 4 | ||
| + | |[[media:Blastn_pSB1K3.nTAT.SOD.his4_premix_29sep.txt|pSB1K3.nTAT.SOD.his4_premix]] | ||
| + | |[[media:Blastn_pSB1K3.nTAT.SOD.his4_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:green">Verified</span>'''. Three silent mutations in His tag; does not alter a.a. sequence. | ||
| + | |- | ||
| + | |pSB1K3.N-TAT⋅SOD⋅His 5 | ||
| + | |[[media:Blastn_pSB1K3.nTAT.SOD.his5_premix_29sep.txt|pSB1K3.nTAT.SOD.his5_premix]] | ||
| + | |[[media:Blastn_pSB1K3.nTAT.SOD.his5_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:green">Verified</span>'''. Three silent mutations in His tag; does not alter a.a. sequence. | ||
| + | |- | ||
| + | |pSB1K3.N-Tra10⋅SOD⋅His 5 | ||
| + | |[[media:PSB1K3.nTra10.SOD.his5_premix_fasta_29sep.txt|pSB1K3.nTra10.SOD.his5_premix]] | ||
| + | |[[media:Blastn_pSB1K3.nTra10.SOD.h5_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:green">Verified</span>'''. Three silent mutations in His tag; does not alter a.a. sequence. | ||
| + | |- | ||
| + | |- | ||
| + | |pSB1K3.N-LMWP⋅SOD⋅His 1 | ||
| + | |[[media:PSB1K3.nLMWP.SOD.his1_premix_fasta_29sep.txt|pSB1K3.nLMWP.SOD.his1_premix]] | ||
| + | |[[media:Blastn_pSB1K3.nLMWP.SOD.hi1_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:green">Verified</span>'''. Three silent mutations in His tag; does not alter a.a. sequence. | ||
| + | |- | ||
| + | |- | ||
| + | |pSB1C3.N-LMWP⋅SOD⋅His 1 | ||
| + | |[[media:PSB1C3.nLMWP.SOD.his1_premix_fasta_29sep.txt|pSB1C3.nLMWP.SOD.his1_premix]] | ||
| + | |[[media:Blastn_pSB1C3.nLMWP.SOD.hi1_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:red">Not verified'''</span>. Incorrect construct. | ||
| + | |- | ||
| + | |pSB1C3.N-LMWP⋅SOD⋅His 4 | ||
| + | |[[media:PSB1C3.nLMWP.SOD.his4_premix_fasta_29sep.txt|pSB1C3.nLMWP.SOD.his4_premix]] | ||
| + | |[[media:Blastn_pSB1C3.nLMWP.SOD.hi4_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:red">Not verified'''</span>. Incorrect construct. | ||
| + | |- | ||
| + | |pEX.SOD | ||
| + | |[[media:PEX.SOD_premix_fasta_29sep.txt|pEX.SOD_premix]] | ||
| + | |[[media:Blastn_pEX.SOD_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:orange">Partly verified'''</span>. Insertion found between SpeI and PstI; does not affect expression. | ||
| + | |- | ||
| + | |pEX.yCCS 5 | ||
| + | |[[media:PEX.yCCS_5_premix_fasta.txt|pEX.yCCS 5_premix]] | ||
| + | |[[media:Blastn_pEX.yCCS_5_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:orange">Partly verified'''</span>. Insertion found between SpeI and PstI; does not affect expression. | ||
| + | |- | ||
| + | |pEX.SOD⋅His | ||
| + | |[[media:PEX.SOD.his_premix_fasta_29sep.txt|pEX.SOD.his_premix]] | ||
| + | |[[media:Blastn_pEX.SOD.his_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:green">Verified'''</span>. Three silent mutations in His tag; does not alter a.a. sequence. | ||
| + | |- | ||
| + | |pEX.His⋅SOD | ||
| + | |[[media:PEX.his.SOD_premix_29sep.txt|pEX.his.SOD_premix]] | ||
| + | |[[media:Blastn_pEX.his.SOD_premix_29sep.txt|Blastn]] | ||
| + | |'''<span style="color:green">Verified'''</span>. Three silent mutations in His tag; does not alter a.a. sequence. | ||
| + | |} | ||
| + | |||
| + | ===Plasmid prep=== | ||
| + | ''From 28/9 ON cultures'' | ||
| + | |||
| + | Spun down ON cultures 10 min, 3000 x ''g''. Decanted supernatant and stored pellets in -20 °C ON for later plasmid prep. | ||
| + | |||
| + | ===Sorting glycerol stocks=== | ||
| + | Started sorting our glycerol stocks into a spreadsheet for future reference. | ||
| + | |||
| + | ==Nina== | ||
| + | |||
| + | ===Agarose gel verificaion=== | ||
| + | |||
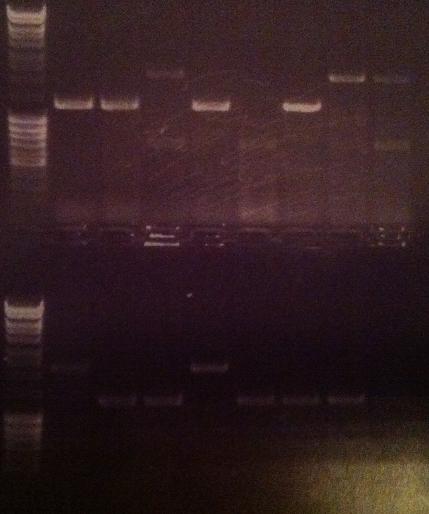
| + | I ran an agarose gel 1 % 100 V on the colony PCR products from yesterday to check if I had any inserts. | ||
| + | |||
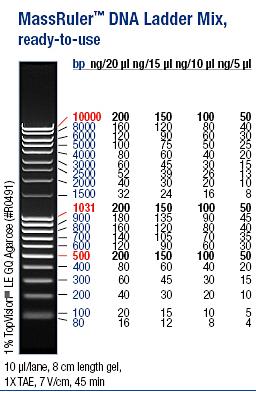
| + | Ladder: MassRuler™ DNA Ladder Mix, ready-to-use, 80-10,000 bp | ||
| + | |||
| + | [[Image:laddermixmassruler.jpg|200px]] | ||
| + | |||
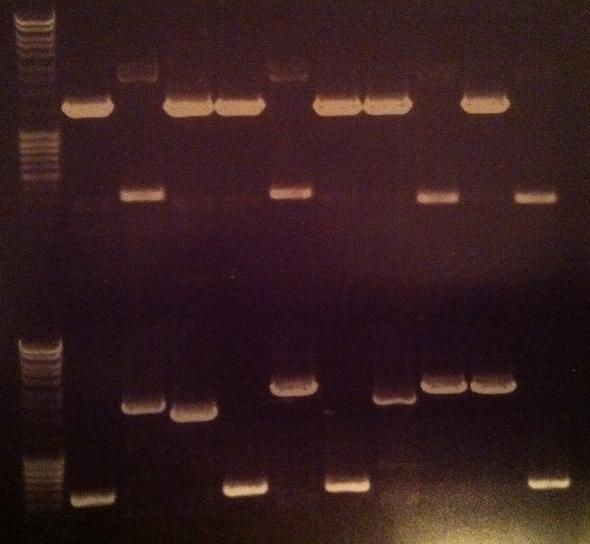
| + | Arrangement on gel: | ||
| + | |||
| + | [[Image:Aq20.jpg]] | ||
| + | |||
| + | From left to right: 1-5 fusion EA, 1-5 Fusion NS, 1-5 IgG LMWP, 1-5 IgG_TAT_N, 1-5 IgG_Tra10_N, 1-5 IgG_TAT_C, 1-5 Protein A_TAT_C, 1-5 pEX, 1-9 pEX | ||
| + | |||
| + | [[Image:Aq21.jpg|200px]] | ||
| + | |||
| + | [[Image:Aq22.jpg|200px]] | ||
| + | |||
| + | [[Image:Aq23.jpg|200px]] | ||
| + | |||
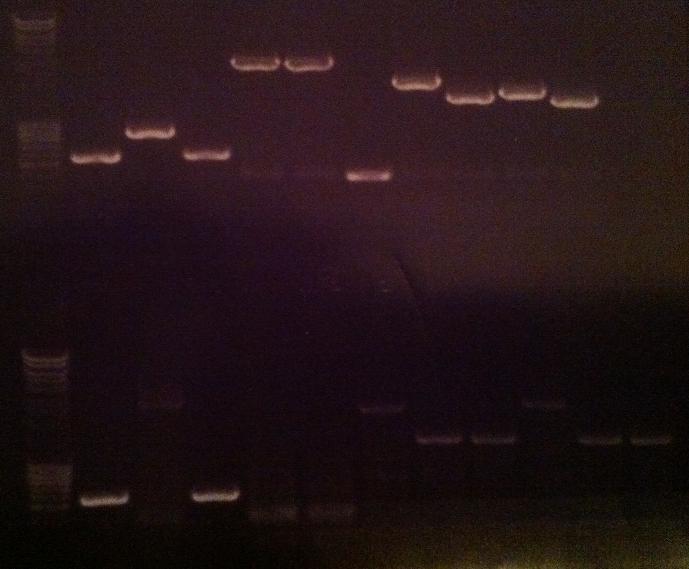
| + | ===Colony PCR=== | ||
| + | |||
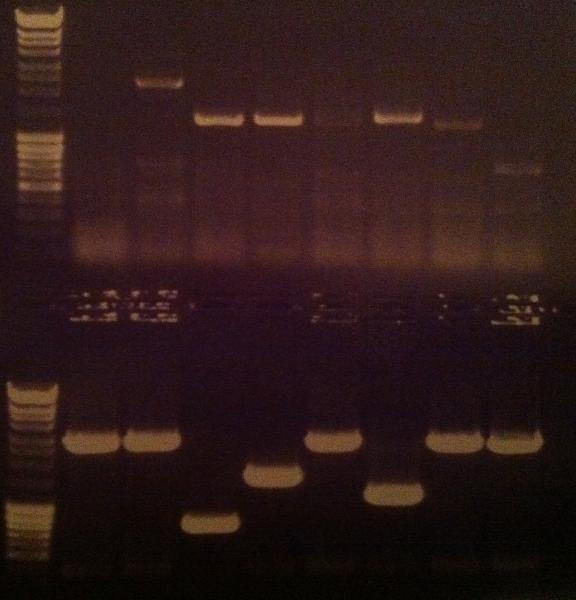
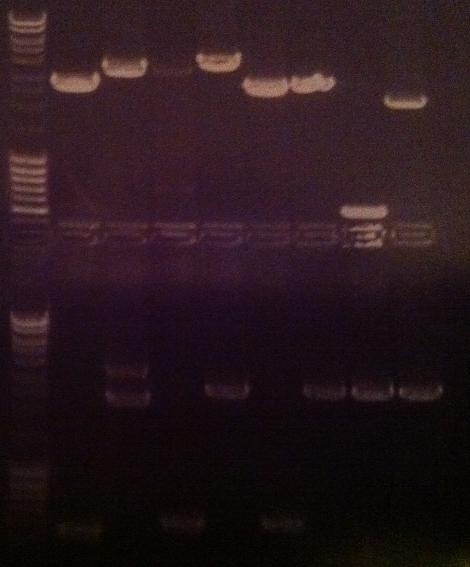
| + | I ran a new colony PCR on the samples I did not have a possitive result on from the gel above. | ||
| + | |||
| + | The samples for this screen are: | ||
| + | |||
| + | IgG_LMWP_N, _TAT_N, _Tra10_N, protein A_LMWP_N_pex, _TAT_N_pex & _Tra10_N_pex | ||
| + | |||
| + | I screened 8 colonies/dish. | ||
| + | |||
| + | PCR master mix: | ||
| + | |||
| + | *MgCl2 8ul | ||
| + | *phusion buffer 5X 80ul | ||
| + | *dNTP 8ul | ||
| + | *primerR 24ul | ||
| + | *primerF 24ul | ||
| + | *polymerase 8ul | ||
| + | *H2O 240ul | ||
| + | |||
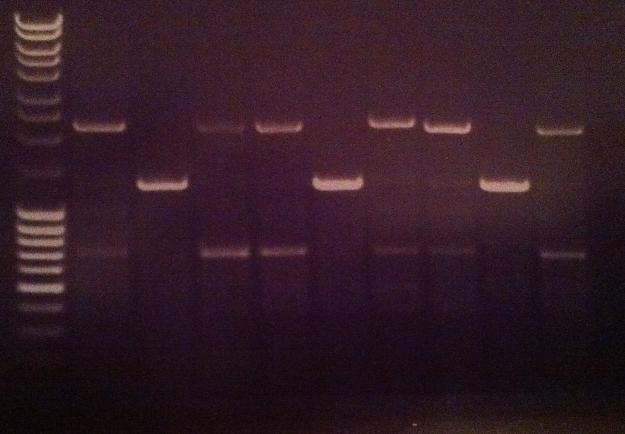
| + | ===Agarose gel verificaion=== | ||
| + | |||
| + | I ran an agarose gel 1 % 100 V on the colony PCR products from yesterday to check if I had any inserts. | ||
| + | |||
| + | Ladder: MassRuler™ DNA Ladder Mix, ready-to-use, 80-10,000 bp | ||
| + | |||
| + | [[Image:laddermixmassruler.jpg|200px]] | ||
| + | |||
| + | Arrangement on gel: | ||
| + | |||
| + | [[Image:Aq24.jpg]] | ||
| + | |||
| + | [[Image:Aq25.jpg|200px]] | ||
| + | |||
| + | [[Image:Aq26.jpg|200px]] | ||
| + | |||
| + | [[Image:Aq27.jpg|200px]] | ||
| + | |||
| + | ===Overnight culture=== | ||
| + | |||
| + | I inoculated Fusion EA colony 1 & 3 and Fusion NS colony 1 & 2 each in 12 ml LB with 24 ul chloramphenicol resistance. | ||
| + | |||
| + | ===Digestion=== | ||
| + | |||
| + | I digested IgG protease and protein A in the bank vector C in order to perform a gel clean up of the genes and insert them into a pMa-His AS vector. | ||
| + | |||
| + | Digestion: | ||
| + | |||
| + | *H2O 15 ul | ||
| + | *DNA 2 ul | ||
| + | *Fastdigest buffer 10X 2 ul | ||
| + | *Restriction enzyme NgoMIV 1 ul | ||
| + | *Restriction enzyme SpeI 1 ul (Add after 1.5h incubation in 37 °C) | ||
| + | |||
| + | |||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === Over expression === | ||
| + | |||
| + | *Load gel | ||
| + | ***dilute 3h samples 1:4 | ||
| + | ***load 8µl in the pre-wells | ||
| + | ***the comb takes up 4µl to load on the gel | ||
| + | ***run PhastGel 20% | ||
| + | |||
| + | {| | ||
| + | ! well | ||
| + | ! sample | ||
| + | | rowspan="6" | [[Image:Place_for_picture.jpg|100px|thumb|left|]] | ||
| + | ! well | ||
| + | ! sample | ||
| + | | rowspan="6" | [[Image:Place_for_picture.jpg|100px|thumb|left|]] | ||
| + | |- | ||
| + | | 1 | ||
| + | | ladder | ||
| + | | 1 | ||
| + | | ladder | ||
| + | |- | ||
| + | | 2 | ||
| + | | SOD 0h | ||
| + | | 2 | ||
| + | | SOD.his 0h | ||
| + | |- | ||
| + | | 3 | ||
| + | | SOD 3h | ||
| + | | 3 | ||
| + | | SOD.his 3h | ||
| + | |- | ||
| + | | 4 | ||
| + | | yCCS 0h | ||
| + | | 4 | ||
| + | | his.SOD 0h | ||
| + | |- | ||
| + | | 5 | ||
| + | | yCCS 3h | ||
| + | | 5 | ||
| + | | his.SOD 3h | ||
| + | |} | ||
| + | |||
| + | ==Johan== | ||
| + | |||
| + | ===Cut CPP-vector=== | ||
| + | |||
| + | 5 µl vector ~1µg | ||
| + | |||
| + | 1 µl NgoMIV | ||
| + | |||
| + | 1 µl EcoRI | ||
| + | |||
| + | 2 µl 10x fastbuffer | ||
| + | |||
| + | 12 µl H2O | ||
| + | |||
| + | Then heat-inactivation of enzymes | ||
| + | |||
| + | ===Cut his-bFGF === | ||
| + | |||
| + | 5 µl his-bFGF (1,5 µl -> 0,3 µg bFGF) | ||
| + | |||
| + | 1 µl AgeI | ||
| + | |||
| + | 1 µl EcoRI | ||
| + | |||
| + | 2 µl 10x fastbuffer | ||
| + | |||
| + | 12 µl H2O | ||
| + | |||
| + | Then heat-inactivation of enzymes | ||
| + | |||
| + | ===Ligation his-bFGF into CPP-vector=== | ||
| + | |||
| + | 5 µl his-bFGF | ||
| + | |||
| + | 0,5 µl cpp-vector | ||
| + | |||
| + | 2 µl buffer | ||
| + | |||
| + | 1 µl T4 ligase | ||
| + | |||
| + | 11,5 µl H2O | ||
| + | |||
| + | ===Transformation=== | ||
| + | |||
| + | 3 µl of all constructs was transformed into top10 cells | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 21:40, 27 October 2010
Contents |
Andreas
Sequencing results
Sequencing results from 27/9 samples arrived and were analyzed by nucleotide BLAST.
| Construct | Sequencing result | Blastn result | Comment |
|---|---|---|---|
| pEX.N-TAT⋅SOD⋅His 3 | pEX.nTAT.SOD.his 3_premix | Blastn | Verified. Three silent mutations in His tag; does not alter a.a. sequence. |
| pEX.N-TAT⋅SOD⋅His 4 | pEX.nTAT.SOD.his 4_premix | Blastn | Not verified. Incorrect construct. |
| pSB1A2.RBS.yCCS 3 | pSB1A2.RBS.yCCS 3_premix | Blastn | Partly verified. Three mutations found: (1) Single base deletion in yCCS sequence, causing a frame shift and an extended open reading frame; (2) Insertion between SpeI and PstI, not affecting expression of cloning; (3) Point mutation in PstI site, disabling PstI restriction. Mutations (1) and (3) are both quite unsure, as the sequencing quality in this area is very low. New sequencing from VR will be sent. |
| pSB1A2.RBS.yCCS 4 | pSB1A2.RBS.yCCS 4_premix | Blastn | Partly verified. Four mutations found: (1) Point mutation changing a K codon to T; (2) Single base deletion in yCCS sequence, causing frame shift; (3) Insertion between SpeI and PstI, not affecting expression of cloning; (4) Point mutation in PstI site, disabling PstI restriction. See comment |
| pSB1K3.N-TAT⋅SOD⋅His 4 | pSB1K3.nTAT.SOD.his4_premix | Blastn | Verified. Three silent mutations in His tag; does not alter a.a. sequence. |
| pSB1K3.N-TAT⋅SOD⋅His 5 | pSB1K3.nTAT.SOD.his5_premix | Blastn | Verified. Three silent mutations in His tag; does not alter a.a. sequence. |
| pSB1K3.N-Tra10⋅SOD⋅His 5 | pSB1K3.nTra10.SOD.his5_premix | Blastn | Verified. Three silent mutations in His tag; does not alter a.a. sequence. |
| pSB1K3.N-LMWP⋅SOD⋅His 1 | pSB1K3.nLMWP.SOD.his1_premix | Blastn | Verified. Three silent mutations in His tag; does not alter a.a. sequence. |
| pSB1C3.N-LMWP⋅SOD⋅His 1 | pSB1C3.nLMWP.SOD.his1_premix | Blastn | Not verified. Incorrect construct. |
| pSB1C3.N-LMWP⋅SOD⋅His 4 | pSB1C3.nLMWP.SOD.his4_premix | Blastn | Not verified. Incorrect construct. |
| pEX.SOD | pEX.SOD_premix | Blastn | Partly verified. Insertion found between SpeI and PstI; does not affect expression. |
| pEX.yCCS 5 | pEX.yCCS 5_premix | Blastn | Partly verified. Insertion found between SpeI and PstI; does not affect expression. |
| pEX.SOD⋅His | pEX.SOD.his_premix | Blastn | Verified. Three silent mutations in His tag; does not alter a.a. sequence. |
| pEX.His⋅SOD | pEX.his.SOD_premix | Blastn | Verified. Three silent mutations in His tag; does not alter a.a. sequence. |
Plasmid prep
From 28/9 ON cultures
Spun down ON cultures 10 min, 3000 x g. Decanted supernatant and stored pellets in -20 °C ON for later plasmid prep.
Sorting glycerol stocks
Started sorting our glycerol stocks into a spreadsheet for future reference.
Nina
Agarose gel verificaion
I ran an agarose gel 1 % 100 V on the colony PCR products from yesterday to check if I had any inserts.
Ladder: MassRuler™ DNA Ladder Mix, ready-to-use, 80-10,000 bp
Arrangement on gel:
From left to right: 1-5 fusion EA, 1-5 Fusion NS, 1-5 IgG LMWP, 1-5 IgG_TAT_N, 1-5 IgG_Tra10_N, 1-5 IgG_TAT_C, 1-5 Protein A_TAT_C, 1-5 pEX, 1-9 pEX
Colony PCR
I ran a new colony PCR on the samples I did not have a possitive result on from the gel above.
The samples for this screen are:
IgG_LMWP_N, _TAT_N, _Tra10_N, protein A_LMWP_N_pex, _TAT_N_pex & _Tra10_N_pex
I screened 8 colonies/dish.
PCR master mix:
- MgCl2 8ul
- phusion buffer 5X 80ul
- dNTP 8ul
- primerR 24ul
- primerF 24ul
- polymerase 8ul
- H2O 240ul
Agarose gel verificaion
I ran an agarose gel 1 % 100 V on the colony PCR products from yesterday to check if I had any inserts.
Ladder: MassRuler™ DNA Ladder Mix, ready-to-use, 80-10,000 bp
Arrangement on gel:
Overnight culture
I inoculated Fusion EA colony 1 & 3 and Fusion NS colony 1 & 2 each in 12 ml LB with 24 ul chloramphenicol resistance.
Digestion
I digested IgG protease and protein A in the bank vector C in order to perform a gel clean up of the genes and insert them into a pMa-His AS vector.
Digestion:
- H2O 15 ul
- DNA 2 ul
- Fastdigest buffer 10X 2 ul
- Restriction enzyme NgoMIV 1 ul
- Restriction enzyme SpeI 1 ul (Add after 1.5h incubation in 37 °C)
Mimmi
Over expression
- Load gel
- dilute 3h samples 1:4
- load 8µl in the pre-wells
- the comb takes up 4µl to load on the gel
- run PhastGel 20%
| well | sample | well | sample | ||
|---|---|---|---|---|---|
| 1 | ladder | 1 | ladder | ||
| 2 | SOD 0h | 2 | SOD.his 0h | ||
| 3 | SOD 3h | 3 | SOD.his 3h | ||
| 4 | yCCS 0h | 4 | his.SOD 0h | ||
| 5 | yCCS 3h | 5 | his.SOD 3h |
Johan
Cut CPP-vector
5 µl vector ~1µg
1 µl NgoMIV
1 µl EcoRI
2 µl 10x fastbuffer
12 µl H2O
Then heat-inactivation of enzymes
Cut his-bFGF
5 µl his-bFGF (1,5 µl -> 0,3 µg bFGF)
1 µl AgeI
1 µl EcoRI
2 µl 10x fastbuffer
12 µl H2O
Then heat-inactivation of enzymes
Ligation his-bFGF into CPP-vector
5 µl his-bFGF
0,5 µl cpp-vector
2 µl buffer
1 µl T4 ligase
11,5 µl H2O
Transformation
3 µl of all constructs was transformed into top10 cells
 |

|
 |

|
 |

|

|

|
 "
"