Team:Stockholm/28 June 2010
From 2010.igem.org
(Difference between revisions)
(New page: {{Stockholm/Top2}} =28 June 2010= = Andreas = ==Colony PCR verification of pEX vector== Colony gradient PCR was run to optimize the colony PCR settings for designed pEX verification pri...) |
m |
||
| (5 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| - | |||
| - | |||
= Andreas = | = Andreas = | ||
| - | ==Colony PCR verification of pEX vector== | + | ===Colony gradient PCR verification of pEX vector=== |
| - | + | Gradient PCR was run to optimize the colony PCR settings for designed pEX verification primers (pEXf and pEXr). Products analyzed by agarose gel electrophoresis. | |
| - | === | + | ====Procedures==== |
| - | == | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | '''PCR settings''' | |
| - | + | * Primers: | |
| - | + | **pEXf forward primer (CGG CTC GTA TAA TGT GTG GAA TTG) | |
| - | + | **pEXr reverse primer (CGT TCA CCG ACA AAC AAC AG) | |
| - | + | * Gradient: 50°C, 55°C, 60°C | |
| - | + | * Elongation time: 2 min 30 s | |
| - | + | ** Expected amplicon size: 116 bp (pEXf) + 103 bp (pEXr) + 1188 bp (insert) = '''1407 bp''''' | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | '''Agarose gel analysis''' | |
| - | + | ||
| - | + | ||
*1% agarose gel | *1% agarose gel | ||
*2 ul sample | *2 ul sample | ||
| - | |||
| - | |||
| - | |||
*170 V, 20 min | *170 V, 20 min | ||
| - | + | ====Results==== | |
'''Figure to be added later.''' | '''Figure to be added later.''' | ||
| - | + | =Hassan= | |
| + | |||
| + | May the "http://www.pathguide.org" guide us through the darkness :) | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | = Mimmi = | ||
| + | |||
| + | === pMA === | ||
| + | |||
| + | ==== Designing primers for verification ==== | ||
| + | |||
| + | *Get vector sequence from iGEM part registry | ||
| + | |||
| + | [[Image:PMA vector.jpg]] | ||
| + | |||
| + | [[Image:PMA vector linear1.jpg]] | ||
| + | |||
| + | *To be able to test in AmplifX, reajust sequence: | ||
| + | |||
| + | [[Image:PMA vector linear2.jpg]] | ||
| + | |||
| + | *In program: Design primers | ||
| + | **primer length | ||
| + | **max Tm difference 2°C | ||
| + | **F primer: 234-284bp | ||
| + | **R primer: 634-684bp | ||
| + | |||
| + | *Choose a primerpair with the same Tm | ||
| + | |||
| + | *"Run PCR" -> looking good! No dimers or unspecific binding | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | === pEX === | ||
| + | |||
| + | ==== Start over night culture for miniprep. ==== | ||
| + | |||
| + | *Mix one colony in 5ml LB containing Amp. (100µg/ml) in a falcon tube | ||
| + | |||
| + | *Leave ON in 37°C, 250rpm with some what opened lid. | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 10:35, 26 October 2010
Contents |
Andreas
Colony gradient PCR verification of pEX vector
Gradient PCR was run to optimize the colony PCR settings for designed pEX verification primers (pEXf and pEXr). Products analyzed by agarose gel electrophoresis.
Procedures
PCR settings
- Primers:
- pEXf forward primer (CGG CTC GTA TAA TGT GTG GAA TTG)
- pEXr reverse primer (CGT TCA CCG ACA AAC AAC AG)
- Gradient: 50°C, 55°C, 60°C
- Elongation time: 2 min 30 s
- Expected amplicon size: 116 bp (pEXf) + 103 bp (pEXr) + 1188 bp (insert) = 1407 bp
Agarose gel analysis
- 1% agarose gel
- 2 ul sample
- 170 V, 20 min
Results
Figure to be added later.
Hassan
May the "http://www.pathguide.org" guide us through the darkness :)
Mimmi
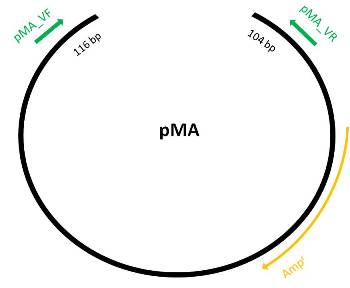
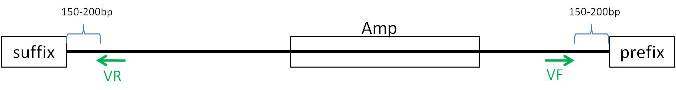
pMA
Designing primers for verification
- Get vector sequence from iGEM part registry
- To be able to test in AmplifX, reajust sequence:
- In program: Design primers
- primer length
- max Tm difference 2°C
- F primer: 234-284bp
- R primer: 634-684bp
- Choose a primerpair with the same Tm
- "Run PCR" -> looking good! No dimers or unspecific binding
pEX
Start over night culture for miniprep.
- Mix one colony in 5ml LB containing Amp. (100µg/ml) in a falcon tube
- Leave ON in 37°C, 250rpm with some what opened lid.
 |

|
 |

|
 |

|

|

|
 "
"