Team:Stockholm/27 August 2010
From 2010.igem.org
(→Hassan) |
(→Andreas) |
||
| Line 21: | Line 21: | ||
==Andreas== | ==Andreas== | ||
| + | ===Troubleshooting of SOD⋅His fusion fail=== | ||
| + | ====Restreak results==== | ||
| + | ''From 26/8'' | ||
| + | |||
| + | All restreaked clones (pSB1K3.SOD⋅His 1 & 2 and pSB1K3.His⋅SOD 1 & 3) were able to grow on both Cm 25 and Km 50. This raises the question whether our plasmid actually carries resistance to both Km and Cm. To test this further, new restreaks were made, where colonies from the Cm 25 plate were streaked on a new Km 50, and vice versa. Also, pSB1K3.RFP, pSB1C3.RFP and untransformed Top10 cells were streaked. | ||
| + | *'''Amp 100 plate''' | ||
| + | **pSB1K3.SOD⋅His 1 from Km 50 plate | ||
| + | **pSB1K3.His⋅SOD 1 from Km 50 plate | ||
| + | **Top10 | ||
| + | **pSB1K3.RFP | ||
| + | **pSB1C3.RFP | ||
| + | *'''Cm 25 plate''' | ||
| + | **pSB1K3.SOD⋅His 1 from Km 50 plate | ||
| + | **pSB1K3.His⋅SOD 1 from Km 50 plate | ||
| + | **Top10 | ||
| + | **pSB1K3.RFP | ||
| + | **pSB1C3.RFP | ||
| + | *'''Km 50 plate''' | ||
| + | **pSB1K3.SOD⋅His 1 from Cm 25 plate | ||
| + | **pSB1K3.His⋅SOD 1 from Cm 25 plate | ||
| + | **Top10 | ||
| + | **pSB1K3.RFP | ||
| + | **pSB1C3.RFP | ||
| + | |||
| + | ===Transfer of SOD into pMA.His (pMA.SOD⋅His)=== | ||
| + | A new strategy attempted for fusing SOD to His. This time SOD will be transfered to pMA carrying the His-tag, thereby avoiding cloning/excising the small tag from its vector. | ||
| + | ====Strategy==== | ||
| + | *pMA.SOD⋅His | ||
| + | **SOD digested with EcoRI and AgeI | ||
| + | **pMA.His digested with EcoRI and NgoMIV | ||
| + | *pMA.His⋅SOD | ||
| + | **SOD digested with NgoMIV and PstI | ||
| + | **pMA.His digested with AgeI and PstI | ||
| + | ====Digestion==== | ||
| + | [pMA.His]=85.67 ng/μl<br /> | ||
| + | [pSB1C3.SOD]=105.5 ng/μl | ||
| + | |||
| + | {|border="1" cellpadding="1" cellspacing="0" | ||
| + | ! | ||
| + | !pMA.His | ||
| + | !pSB1C3.SOD | ||
| + | |- | ||
| + | |10X FD buffer | ||
| + | |align="center"|2 | ||
| + | |align="center"|2 | ||
| + | |- | ||
| + | |DNA (1 μg) | ||
| + | |align="center"|11.7 | ||
| + | |align="center"|9.5 | ||
| + | |- | ||
| + | |dH<sub>2</sub>O | ||
| + | |align="center"|4.3 | ||
| + | |align="center"|6.5 | ||
| + | |- | ||
| + | |FD EcoRI | ||
| + | |align="center"|1 | ||
| + | |align="center"|1 | ||
| + | |- | ||
| + | |NgoMIV | ||
| + | |align="center"|1 | ||
| + | |align="center"|0 | ||
| + | |- | ||
| + | |FD AgeI | ||
| + | |align="center"|0 | ||
| + | |align="center"|1 | ||
| + | |- | ||
| + | | | ||
| + | !20 | ||
| + | !20 | ||
| + | |} | ||
| + | |||
| + | '''Incubation''' | ||
| + | *'''FastDigest enzymes:''' 37 °C, 30 min | ||
| + | *'''NgoMIV:''' 37 °C, 2 h | ||
| + | |||
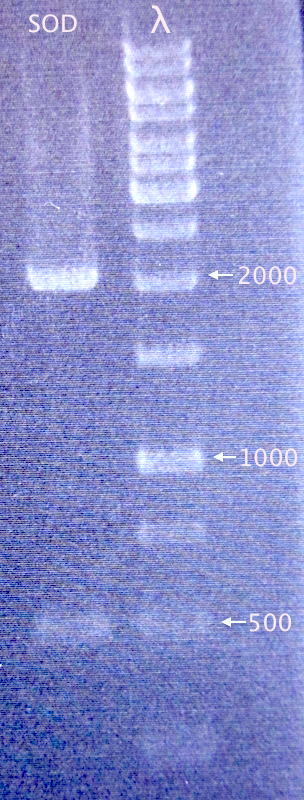
| + | =====Gel verification===== | ||
| + | [[image:Gelver_dig_SOD_27aug.png|150px|right|thumb|'''Gel verification of SOD digested with EcoRI and AgeI.'''<br />3 μl λ; 3 μl sample.<br />λ=O'GeneRuler 1 kb DNA ladder.]] | ||
| + | *Dig.SOD E+N (SOD) | ||
| + | |||
| + | 1% agarose, 100 V, 45 min | ||
| + | |||
| + | '''Results'''<br /> | ||
| + | Gel shows successful digestion: no undigested plasmid and a band corresponding well to the expected SOD size (492 bp). | ||
| + | |||
| + | ====Gel extraction==== | ||
| + | Remaining 22 μl SOD sample run on 1 % agarose gel, 100 V. SOD DNA band excised (240 mg gel; "Extr. Dig SOD E+A") and saved in -20 °C until tomorrow. | ||
| + | |||
| + | ===Isolation of CPPs from N-CPP cluster=== | ||
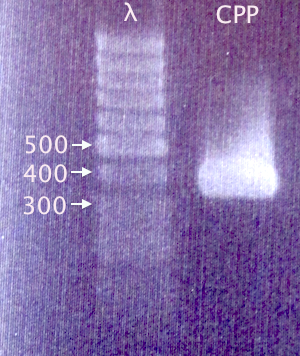
| + | ====Gel verification==== | ||
| + | [[image:Gelver_CPPcluster_PCR_27aug.png|180px|right|thumb|'''Gel verification of CPP cluster PCR.'''<br />3 μl λ; 3 μl sample.<br />λ=O'GeneRuler 1 kb DNA ladder.]] | ||
| + | Gel verification of Nina's CPP cluster PCR | ||
| + | |||
| + | 1.5 % agarose, 100 V, 25 min<br /> | ||
| + | '''Expected band:''' 379 bp | ||
| + | |||
| + | ====Gel extraction==== | ||
| + | ≈ 35 μl sample. CPP cluster band excised (220 mg gel; "Extr. N-CPP clust") and saved in -20 °C. | ||
Revision as of 10:21, 30 August 2010
Contents |
Hassan
added some text, animation is also a little slower now, a border added.
other things to be changed now are:
- a darker color for after pigmentation.
- colors need to be changed for proteins and genes.
- a more proper timing for after pigmentation.
Andreas
Troubleshooting of SOD⋅His fusion fail
Restreak results
From 26/8
All restreaked clones (pSB1K3.SOD⋅His 1 & 2 and pSB1K3.His⋅SOD 1 & 3) were able to grow on both Cm 25 and Km 50. This raises the question whether our plasmid actually carries resistance to both Km and Cm. To test this further, new restreaks were made, where colonies from the Cm 25 plate were streaked on a new Km 50, and vice versa. Also, pSB1K3.RFP, pSB1C3.RFP and untransformed Top10 cells were streaked.
- Amp 100 plate
- pSB1K3.SOD⋅His 1 from Km 50 plate
- pSB1K3.His⋅SOD 1 from Km 50 plate
- Top10
- pSB1K3.RFP
- pSB1C3.RFP
- Cm 25 plate
- pSB1K3.SOD⋅His 1 from Km 50 plate
- pSB1K3.His⋅SOD 1 from Km 50 plate
- Top10
- pSB1K3.RFP
- pSB1C3.RFP
- Km 50 plate
- pSB1K3.SOD⋅His 1 from Cm 25 plate
- pSB1K3.His⋅SOD 1 from Cm 25 plate
- Top10
- pSB1K3.RFP
- pSB1C3.RFP
Transfer of SOD into pMA.His (pMA.SOD⋅His)
A new strategy attempted for fusing SOD to His. This time SOD will be transfered to pMA carrying the His-tag, thereby avoiding cloning/excising the small tag from its vector.
Strategy
- pMA.SOD⋅His
- SOD digested with EcoRI and AgeI
- pMA.His digested with EcoRI and NgoMIV
- pMA.His⋅SOD
- SOD digested with NgoMIV and PstI
- pMA.His digested with AgeI and PstI
Digestion
[pMA.His]=85.67 ng/μl
[pSB1C3.SOD]=105.5 ng/μl
| pMA.His | pSB1C3.SOD | |
|---|---|---|
| 10X FD buffer | 2 | 2 |
| DNA (1 μg) | 11.7 | 9.5 |
| dH2O | 4.3 | 6.5 |
| FD EcoRI | 1 | 1 |
| NgoMIV | 1 | 0 |
| FD AgeI | 0 | 1 |
| 20 | 20 |
Incubation
- FastDigest enzymes: 37 °C, 30 min
- NgoMIV: 37 °C, 2 h
Gel verification
- Dig.SOD E+N (SOD)
1% agarose, 100 V, 45 min
Results
Gel shows successful digestion: no undigested plasmid and a band corresponding well to the expected SOD size (492 bp).
Gel extraction
Remaining 22 μl SOD sample run on 1 % agarose gel, 100 V. SOD DNA band excised (240 mg gel; "Extr. Dig SOD E+A") and saved in -20 °C until tomorrow.
Isolation of CPPs from N-CPP cluster
Gel verification
Gel verification of Nina's CPP cluster PCR
1.5 % agarose, 100 V, 25 min
Expected band: 379 bp
Gel extraction
≈ 35 μl sample. CPP cluster band excised (220 mg gel; "Extr. N-CPP clust") and saved in -20 °C.
 "
"