Team:Stockholm/23 August 2010
From 2010.igem.org
Contents |
Andreas
Assembly of SOD/yCCS⋅His constructs into pSB1K3
Continued from 21/8
Plasmid prep
| DNA concentrations | ||||
|---|---|---|---|---|
| Before sample conc. | After sample conc. | |||
| Sample | Conc. [ng/μl] | A260/A280 | Conc. [ng/μl] | A260/A280 |
| pSB1K3.SOD⋅His 1† | 61.50 | 1.86 | 100.9 | 1.80 |
| pSB1K3.SOD⋅His 2† | 76.45 | 1.78 | 126.1 | 1.83 |
| pSB1K3.yCCS⋅His 2 | 171.5 | 1.88 | – | – |
| pSB1K3.yCSS⋅His 3† | 84.15 | 1.88 | 133.6 | 1.84 |
From 21/8 cultures
- E.Z.N.A Plasmid Mini kit I
- 50 μl elution volume
Sequencing
Samples prepared for sequencing:
- 15 μl DNA (>100 ng/μl)
- 1.5 μl 10 μM pSB-VR primer
| Sample | Label | Seq # |
|---|---|---|
| pSB1K3.SOD⋅His 1 | pK-SH1 | 938 |
| pSB1K3.SOD⋅His 2 | pK-SH2 | 939 |
| pSB1K3.yCCS⋅His 2 | pK-yH2 | 940 |
| pSB1K3.yCCS⋅His 3 | pK-yH3 | 941 |
Enzyme inactivation
Inactivated restriction enzymes in digestion samples from 19/8 and 20/8 in 80 °C, 10 min.
- Dig pEX X+P
- Dig pC.RFP X+P
- Dig His E+A
- Dig m-yCCS N+P
- Dig m-SOD N+P
Transfer of RFP coding device to pEX
Colony restreaks
Results from 21/8
All clones grew well on the Amp plate, but not on Km, indicating RFP has indeed been transfered from pSB1K3 to a target AmpR vector, and that pSB1K3 does not express AmpR.
I later realized that the transfer of RFP was not from pSB1K3, but from pSB1C3, making this restreak pointless.
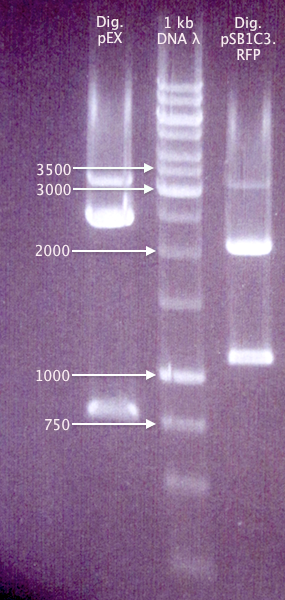
Gel verification of Dig pEX X+P and Dig pC.RFP X+P
Ran a gel of the digestion samples from 19/8 to verify the sizes of vectors and inserts.
1 % agarose, 100 V, 1 h 20 min
Expected bands
- Dig pEX X+P
- 4453 bp (vector)
- 1237 bp (insert)
- Dig pC.RFP X+P
- 2054 bp (pSB1C3 vector)
- 1095 bp (RFP insert)
Results
The bands for "pC.RFP X+P" correspond well to what was expected. However, none of the "pEX X+P" bands do. It might be possible that this is actually a pMA vector, carrying a ≈800 bp insert (unclear what). This would explain why ligations and transformations from 19/8 resulted in red colonies, but colony PCR amplification with pEX primers doesn't result in any bands.
Digestion of new pEX vector
Used pEX vector prepared and verified 21/8 for new pEX digestion with XbaI and PstI.
| Digestion mix | ||
|---|---|---|
| 10X FD buffer | 3 μl | [DNA] = 33.3 ng/μl Inc.: 37 °C, 30 min |
| dH2O | 7 μl | |
| 1 μg DNA (pEX) | 18 μl | |
| FD XbaI | 1 μl | |
| FD PstI | 1 μl | |
| 30 μl | ||
- after
Gel verification
1 % agarose, 100 V, 35 min
Results
pEX confirmed - bands correspond well.
Ligation
| Ligation mix | |||
|---|---|---|---|
| 100 ng vector (pEX) | 3 μl | 1/3 ratio | [Dig pEX X+P] = 33.3 ng/μl (23/8) [Dig pC.RFP X+P]=66.6 ng/μl (19/8) Inc.: 22 °C, 10 min |
| 165 ng insert (RFP) | 2.5 μl | ||
| 5X Rapid Ligation buf. | 4 μl | ||
| dH2O | 9.5 μl | ||
| T4 DNA Ligase | 1 μl | ||
| 20 μl | |||
Transformation
Procedures based on quick transformation protocol:
- 1.5 μl ligation mix. 15 min incubation on ice.
- 30 s heat-shock in 42 °C
Plating onto 100 Amp LB agar. 37 °C ON.
ON cultures
Set ON cultures for preparation of glycerol stocks:
- 3 ml LB + Km 50:
- pSB1K3.SOD⋅His 1
- pSB1K3.SOD⋅His 2
- pSB1K3.yCCS⋅His 2
- pSB1K3.yCCS⋅His 3
- 3 ml LB + Amp 100:
- pMA.His (Picked 19/8)
- pEX (Picked 19/8)
Inc. 30 °C, ON.
Assembly of His⋅SOD/yCCS constructs
Ligation
Used digestion samples from 20/8 for ligations. Ligation protocol identical to that from 20/8.
Transformation
Standard transformation protocol.
- 1 μl ligation mix
- Plating on Km 50.
 "
"