Team:Stockholm/22 September 2010
From 2010.igem.org
(Difference between revisions)
(→Andreas) |
|||
| Line 29: | Line 29: | ||
::'''***Note to self***'''<br /> | ::'''***Note to self***'''<br /> | ||
::''It might be possible that pSB1A2.RBS⋅yCCS clones have been mixed up with pEX.N-TAT⋅SOD⋅His. After plasmid prep, this should be tested by digesting the samples and analyze digestion sizes. | ::''It might be possible that pSB1A2.RBS⋅yCCS clones have been mixed up with pEX.N-TAT⋅SOD⋅His. After plasmid prep, this should be tested by digesting the samples and analyze digestion sizes. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === SOD / SOD.his / his.SOD / yCCS === | ||
| + | |||
| + | {| | ||
| + | | colspan="3" | ''colony PCR'' | ||
| + | | rowspan="8" width="100" | | ||
| + | ! samples | ||
| + | | rowspan="8" width="100" | | ||
| + | ! colspan="2" | Conditions | ||
| + | | rowspan="3" | | ||
| + | |- | ||
| + | | colspan="3" | | ||
| + | | pEX.SOD x3 | ||
| + | ! time | ||
| + | ! °C | ||
| + | |- | ||
| + | ! mix | ||
| + | | (µl) | ||
| + | | x13 | ||
| + | | pEX.SOD.his x3 | ||
| + | | 5m | ||
| + | | 95 | ||
| + | |- | ||
| + | | mastermix | ||
| + | | 24.5 | ||
| + | | rowspan="5" | | ||
| + | | pEX.his.SOD x3 | ||
| + | | 30s | ||
| + | | 95 | ||
| + | | ) | ||
| + | |- | ||
| + | | DNA | ||
| + | | 0.5 | ||
| + | | pEX.yCCS x3 | ||
| + | | 30s | ||
| + | | 55 | ||
| + | | > 30 cycles | ||
| + | |- | ||
| + | | align="right" | tot | ||
| + | | 25µl | ||
| + | | rowspan="3" | | ||
| + | | 1m20s | ||
| + | | 72 | ||
| + | | ) | ||
| + | |- | ||
| + | | colspan="2" rowspan="2" | | ||
| + | | 5m | ||
| + | | 72 | ||
| + | |- | ||
| + | | oo | ||
| + | | 25 | ||
| + | |} | ||
| + | |||
| + | |||
| + | |||
| + | ==== Gel ==== | ||
| + | |||
| + | [[Image:place_for_picture.jpg|200px|thumb|left|]] | ||
| + | {| | ||
| + | ! well | ||
| + | ! width="120" | sample | ||
| + | ! well | ||
| + | ! sample | ||
| + | |- | ||
| + | | 1 | ||
| + | | ladder | ||
| + | | 9 | ||
| + | | ladder | ||
| + | |- | ||
| + | | 2 | ||
| + | | pEX.SOD.his 1 | ||
| + | | 10 | ||
| + | | pEX.SOD 1 | ||
| + | |- | ||
| + | | 3 | ||
| + | | pEX.SOD.his 2 | ||
| + | | 11 | ||
| + | | pEX.SOD 2 | ||
| + | |- | ||
| + | | 4 | ||
| + | | pEX.SOD.his 3 | ||
| + | | 12 | ||
| + | | pEX.SOD 3 | ||
| + | |- | ||
| + | | 5 | ||
| + | | pEX.his.SOD 1 | ||
| + | | 13 | ||
| + | | pEX.yCCS 1 | ||
| + | |- | ||
| + | | 6 | ||
| + | | pEX.his.SOD 1 | ||
| + | | 14 | ||
| + | | pEX.yCCS 2 | ||
| + | |- | ||
| + | | 7 | ||
| + | | pEX.his.SOD 1 | ||
| + | | 15 | ||
| + | | pEX.yCCS 2 | ||
| + | |- | ||
| + | | 8 | ||
| + | | pEX.SOD.his | ||
| + | | 16 | ||
| + | | - | ||
| + | |} | ||
Revision as of 13:01, 4 October 2010
Contents |
Andreas
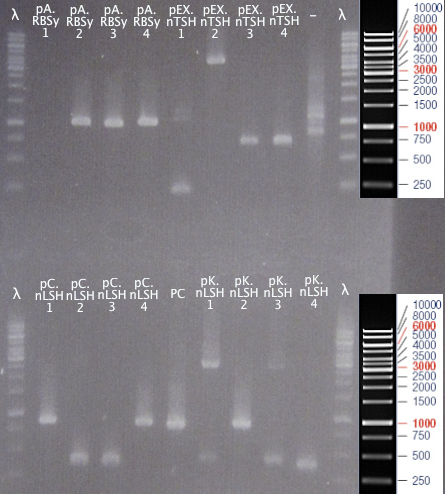
Gel verification
Of 21/9 colony PCR samples
1 % agarose, 120 V
Expected bands
- pSB1A2.RBS⋅yCCS (pA.RBSy): 1018 bp
- pEX.N-TAT⋅SOD⋅His (pEX.nTSH): 735 bp
- pSB1C3.N-LMWP⋅SOD⋅His (pC.nLSH): 857 bp
- pSB1K3.N-LMWP⋅SOD⋅His (pK.nLSH): 857 bp
- pSB1C3.SOD⋅His (PC): 815 bp
Results
- pSB1A2.RBS⋅yCCS: Seemingly correct-sized bands for clones 2, 3 and 4.
- pEX.N-TAT⋅SOD⋅His: Seemingly correct-sized bands for clones 3 & 4.
- pSB1C3.N-LMWP⋅SOD⋅His: Seemingly correct-sized bands for clones 1 & 4.
- pSB1K3.N-LMWP⋅SOD⋅His: Seemingly correct-sized band for clone 1.
ON cultures
- 5 ml LB + appr. antibiotic (100 Amp, 25 Cm or 50 Km); 37 °C, 225 rpm
- pSB1A2.RBS⋅yCCS: clones 3 and 4
- pEX.N-TAT⋅SOD⋅His: clones 3 and 4
- pSB1C3.N-LMWP⋅SOD⋅His: clones 1 and 4
- pSB1K3.N-LMWP⋅SOD⋅His: clone 1
- ***Note to self***
- It might be possible that pSB1A2.RBS⋅yCCS clones have been mixed up with pEX.N-TAT⋅SOD⋅His. After plasmid prep, this should be tested by digesting the samples and analyze digestion sizes.
- ***Note to self***
Mimmi
SOD / SOD.his / his.SOD / yCCS
| colony PCR | samples | Conditions | ||||||
|---|---|---|---|---|---|---|---|---|
| pEX.SOD x3 | time | °C | ||||||
| mix | (µl) | x13 | pEX.SOD.his x3 | 5m | 95 | |||
| mastermix | 24.5 | pEX.his.SOD x3 | 30s | 95 | ) | |||
| DNA | 0.5 | pEX.yCCS x3 | 30s | 55 | > 30 cycles | |||
| tot | 25µl | 1m20s | 72 | ) | ||||
| 5m | 72 | |||||||
| oo | 25 | |||||||
Gel
| well | sample | well | sample |
|---|---|---|---|
| 1 | ladder | 9 | ladder |
| 2 | pEX.SOD.his 1 | 10 | pEX.SOD 1 |
| 3 | pEX.SOD.his 2 | 11 | pEX.SOD 2 |
| 4 | pEX.SOD.his 3 | 12 | pEX.SOD 3 |
| 5 | pEX.his.SOD 1 | 13 | pEX.yCCS 1 |
| 6 | pEX.his.SOD 1 | 14 | pEX.yCCS 2 |
| 7 | pEX.his.SOD 1 | 15 | pEX.yCCS 2 |
| 8 | pEX.SOD.his | 16 | - |
 "
"