Team:Stockholm/16 September 2010
From 2010.igem.org
(→N-CPP sequencing) |
(→Andreas) |
||
| Line 81: | Line 81: | ||
*pSB1C3.N-Tra10: clone 5 ([[media:Blastn_pSB.N-Tra10_pSB1C3.nCPP_15sep.txt|Blastn]]) | *pSB1C3.N-Tra10: clone 5 ([[media:Blastn_pSB.N-Tra10_pSB1C3.nCPP_15sep.txt|Blastn]]) | ||
*pSB1C3.N-LMWP: clones 2, 3 & 11 ([[media:Blastn_pSB.N-LMWP_pSB1C3.nCPP_15sep.txt|Blastn]]) | *pSB1C3.N-LMWP: clones 2, 3 & 11 ([[media:Blastn_pSB.N-LMWP_pSB1C3.nCPP_15sep.txt|Blastn]]) | ||
| + | |||
| + | ====ON cultures==== | ||
| + | Set ON cultures for plasmid prep (5 ml LB + 25 Cm; 37 °C, 220 rpm) and glycerol stocks (3 ml LB + 25 Cm; 30 °C). | ||
| + | *Clone 5: pSB1C3.N-Tra10 | ||
| + | *Clone 11: pSB1C3.N-LMWP | ||
| + | *Clone 12: pSB1C3.N-TAT | ||
Revision as of 21:21, 21 September 2010
Contents |
Andreas
Assembly of new parts
Gel verification of part digestions
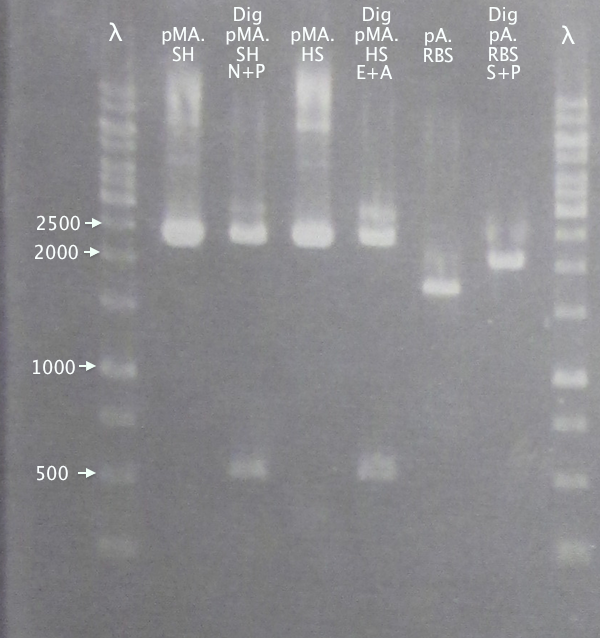
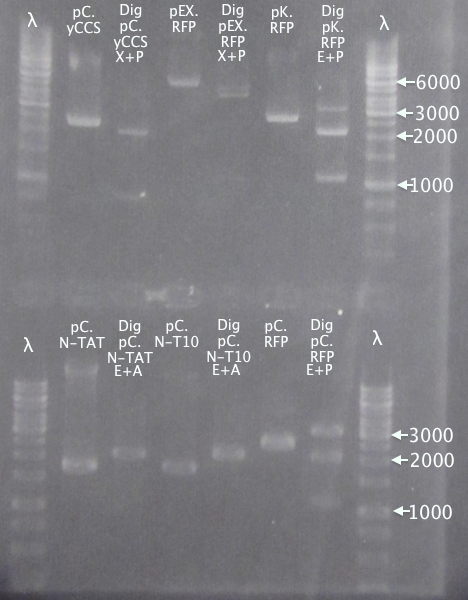
Ran gels of digestion samples in parallel with undigested samples to verify successful digestions and insert sizes.
Gel 1
1 % agarose, 110 V
Gel 2
1 % agarose, 110 V
Results
Successful digestion with corresponding bands for all digested samples. N-TAT and N-Tra10 not verified, since gel was run too far, but plasmid linearization should indicate successful digestion.
Most samples show somewhat incomplete digestion. For digestions with FastDigest enzymes, this may be an indication of old/inactive enzymes. Especially PstI should be analyzed for activity.
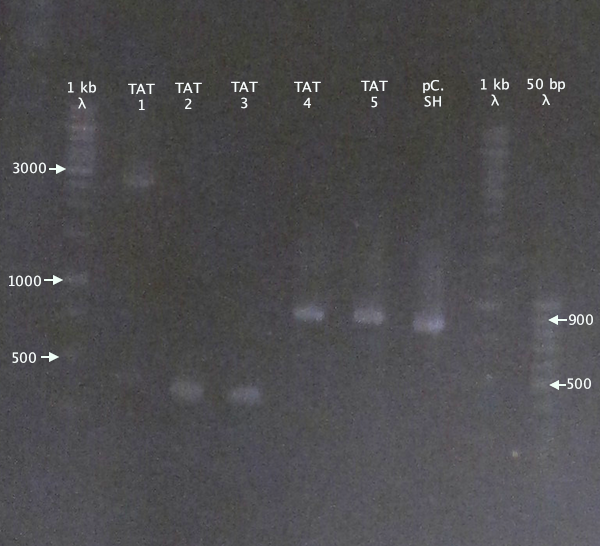
Colony PCR
Picked new colonies for colony PCR from 14/9 plates:
- pSB1K3.N-TAT⋅SOD⋅His: TAT⋅SH 1-5
- pSB1K3.N-Tra10⋅SOD⋅His: Tra10⋅SH 1-5
- pEX.SOD 1-4
Standard colony PCR settings.
- Elongation: 1:30
Gel verification
Gel 1
1 % agarose, 110 V
Expected bands
- pSB1K3.N-TAT⋅SOD⋅His (TAT): 848 bp
- pSB1C3.SOD⋅His (pC.SH): 815 bp
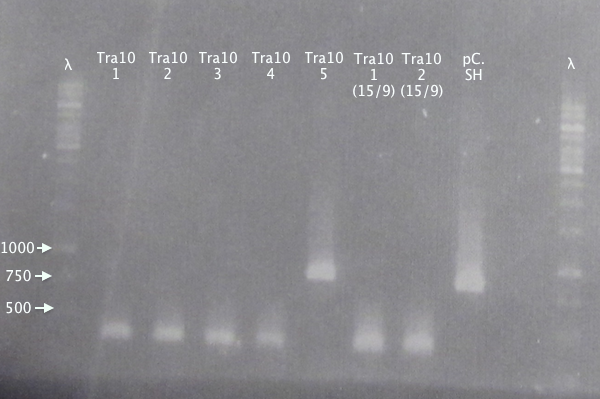
Gel 2
1 % agarose, 110 V
Expected bands
- pSB1K3.N-Tra10⋅SOD⋅His (Tra10): 878 bp
- pSB1C3.SOD⋅His (pC.SH): 815 bp
Gel 3
1 % agarose, 110 V
Expected bands
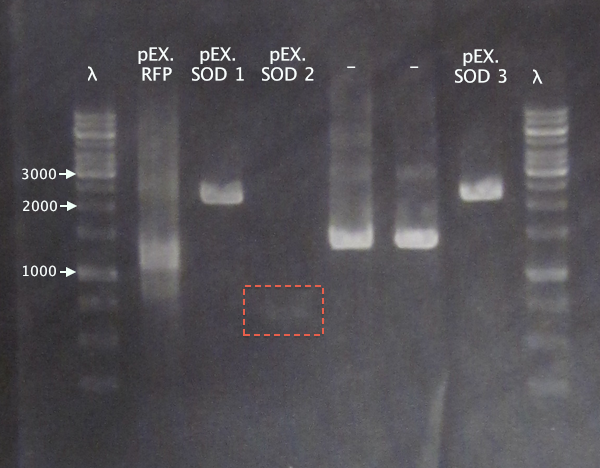
- pEX.SOD: 678 bp
- pEX.RFP: 862 bp, 1010 bp, 1124 bp, 1272 bp
Results
pSB1K3.N-TAT⋅SOD⋅His clones 4 and 5 seem slightly larger than the control, pSB1C3.SOD⋅His, indicating correct assembly.
Of the pSB1K3.N-Tra10⋅SOD⋅His samples, clone 5 seems correct, compared to the control (same as above).
For the pEX.SOD samples the results are very strange. Two of the clones resulted in way too large bands (≈2500 bp); unclear what these vectors carry. Clone 2 resulted in a very weak band with the correct size. This clone was chosen for ON growth.
ON cultures
Set ON cultures (5 ml LB + 50 Km or 100 Amp; 37 °C, 220 rpm) of the following:
- pSB1K3.N-TAT⋅SOD⋅His 4
- pSB1K3.N-TAT⋅SOD⋅His 5
- pSB1K3.N-Tra10⋅SOD⋅His 5
- pEX.SOD 2
N-CPP sequencing
Sequencing results from 13/9 returned.
- pSB1C3.nCCP 2 (fasta)
- pSB1C3.nCCP 3 (fasta)
- pSB1C3.nCCP 5 (fasta)
- pSB1C3.nCCP 8 (fasta)
- pSB1C3.nCCP 9 (fasta)
- pSB1C3.nCCP 10 (fasta)
- pSB1C3.nCCP 11 (fasta)
- pSB1C3.nCCP 12 (fasta)
Ran multiple nucleotide Blast (Blastn) alignments to identify the three N-CPPs from the sequence:
- pSB1C3.N-TAT: clones 9 & 12 (Blastn)
- pSB1C3.N-Tra10: clone 5 (Blastn)
- pSB1C3.N-LMWP: clones 2, 3 & 11 (Blastn)
ON cultures
Set ON cultures for plasmid prep (5 ml LB + 25 Cm; 37 °C, 220 rpm) and glycerol stocks (3 ml LB + 25 Cm; 30 °C).
- Clone 5: pSB1C3.N-Tra10
- Clone 11: pSB1C3.N-LMWP
- Clone 12: pSB1C3.N-TAT
 "
"