Team:Stockholm/12 October 2010
From 2010.igem.org
(→Andreas) |
|||
| (4 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Stockholm/Top2}} | {{Stockholm/Top2}} | ||
| + | ==Nina== | ||
| + | |||
| + | ===Gel clean up=== | ||
| + | |||
| + | I loaded samples in an agarose gel 1 % and ran at 80 V. | ||
| + | |||
| + | Samples: | ||
| + | |||
| + | *Protein A_TAT_N, _Tra10_N and _LMWP_N | ||
| + | *Fusion_CPP1, TAT_C ans _CPP3 | ||
| + | *Fusion_TAT_N, _Tra10_N and _LMWP_N | ||
| + | |||
| + | The procedure was according to the description in protocols. | ||
| + | |||
| + | Weight of all samples: 200 mg | ||
| + | |||
| + | 200 * 3 = 600 ul QXI | ||
| + | |||
| + | In step 3 I added 10 ul of QIAEXII to each sample. | ||
| + | |||
| + | ===Polyacrylamide gel=== | ||
| + | |||
| + | I ran a gel of the purified SOD.His (N terminal) to check if there had been any purification of the protein. | ||
| + | |||
| + | Ladder: PageRuler Unst. Protein Ladder | ||
| + | |||
| + | Arrangement on the gel: | ||
| + | |||
| + | [[Image:Aq18.jpg]] | ||
| + | |||
| + | Gel: | ||
| + | |||
| + | [[Image:Aq19.jpg|300px]] | ||
| + | |||
| + | The gel was incubated in coomassie blue staining and destained on shake. | ||
| + | |||
| + | Unfortunately there were no bands representing SOD.His (~17 kDa) on the gel, which must mean that there was too small culture (12 ml) and that the cells have not been lysed properly. I will redo this work but with more start culture and lyse the cells in a more effiecient way. | ||
| + | |||
| + | |||
| + | The polyacrylamide gel was prepared by the SDS-PAGE mixtures described in protocols. | ||
| + | |||
==Andreas== | ==Andreas== | ||
===Removal of insertion in BioBrick suffixes=== | ===Removal of insertion in BioBrick suffixes=== | ||
| Line 36: | Line 77: | ||
#Correct size for clone C, weak band for clone B. Clone A probably carrying an RFP insert. | #Correct size for clone C, weak band for clone B. Clone A probably carrying an RFP insert. | ||
#All clones correct. | #All clones correct. | ||
| + | |||
| + | ====ON cultures==== | ||
| + | *5 ml LB + Cm 25; 37 °C, 225 rpm | ||
| + | **1C, 2C, 3C, 4C, 5C | ||
| + | |||
| + | ===BL21 transformation and growth test=== | ||
| + | Testing BL21 cells with/without CPP expression by transforming and inducing with IPTG. | ||
| + | |||
| + | *50 μl 0.1 M IPTG | ||
| + | *40 μl competent cells | ||
| + | **pEX.SOD⋅His | ||
| + | **pEX.nTra10⋅SOD⋅His | ||
| + | **pEX.nTAT⋅SOD⋅His | ||
| + | **pEX.nLMWP⋅SOD⋅His | ||
| + | |||
| + | |||
| + | |||
| + | == Mimmi == | ||
| + | |||
| + | === SOD activity assay === | ||
| + | |||
| + | *Kits? | ||
| + | **Biosite/trevigen 6000sek | ||
| + | **Biovision/labinova 3000sek, ca 1week delivery time | ||
| + | **''Cayman'' | ||
| + | **''Cellbiolabs'' | ||
| + | **''Merch'' | ||
| + | **Abcam 3600sek | ||
| + | **Probior | ||
| + | **Sigma-Aldrich 3000sek | ||
| + | **''T-cell technology inc.'' | ||
| + | |||
| + | |||
| + | |||
| + | *Kit from Sigma-Aldrich | ||
| + | **sponsored with a very good offer | ||
| + | |||
| + | {{Stockholm/Footer}} | ||
Latest revision as of 11:09, 26 October 2010
Contents |
Nina
Gel clean up
I loaded samples in an agarose gel 1 % and ran at 80 V.
Samples:
- Protein A_TAT_N, _Tra10_N and _LMWP_N
- Fusion_CPP1, TAT_C ans _CPP3
- Fusion_TAT_N, _Tra10_N and _LMWP_N
The procedure was according to the description in protocols.
Weight of all samples: 200 mg
200 * 3 = 600 ul QXI
In step 3 I added 10 ul of QIAEXII to each sample.
Polyacrylamide gel
I ran a gel of the purified SOD.His (N terminal) to check if there had been any purification of the protein.
Ladder: PageRuler Unst. Protein Ladder
Arrangement on the gel:
Gel:
The gel was incubated in coomassie blue staining and destained on shake.
Unfortunately there were no bands representing SOD.His (~17 kDa) on the gel, which must mean that there was too small culture (12 ml) and that the cells have not been lysed properly. I will redo this work but with more start culture and lyse the cells in a more effiecient way.
The polyacrylamide gel was prepared by the SDS-PAGE mixtures described in protocols.
Andreas
Removal of insertion in BioBrick suffixes
Transformation results
Good colony yield on all plates. A few red colonies discovered on all plates, indicating that some partly digested pSB1C3 vector, still containing the RFP insert, had been excised during gel extraction.
Colony PCR
From 11/10 transformations'
- pSB1C3.IgGp A-C
- pSB1C3.bFGF A-C
- pSB1C3.ProtA A-C
- pSB1C3.yCCS A-C
- pSB1C3.SOD A-C
- Standard colony PCR settings
- 1:15 elongation time
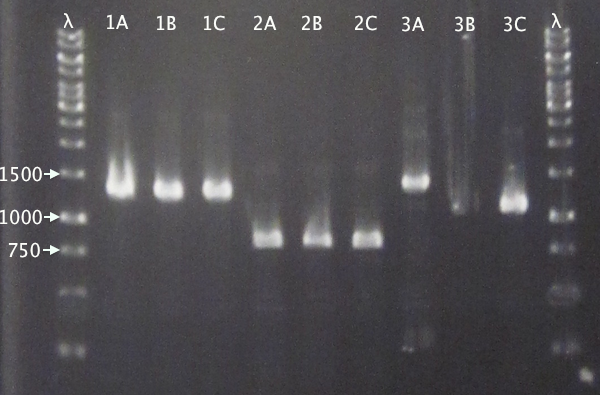
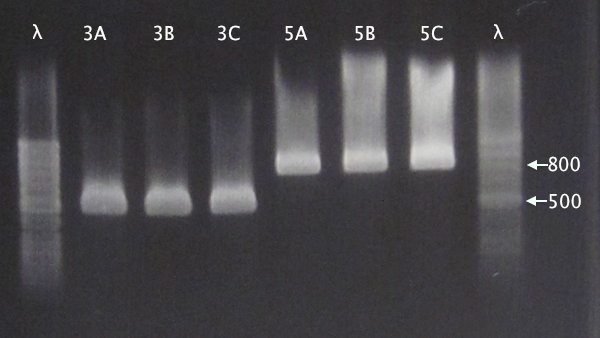
Gel verification
Gel 1: 1 % agarose, 140 V
Gel 2: 1 % agarose, 160 V
Expected bands
- pSB1C3.IgGp: 1262 bp
- pSB1C3.bFGF: 794 bp
- pSB1C3.ProtA: 506 bp
- pSB1C3.yCCS: 1076 bp
- pSB1C3.SOD: 791 bp
Results
- All clones correct.
- All clones correct.
- All clones correct.
- Correct size for clone C, weak band for clone B. Clone A probably carrying an RFP insert.
- All clones correct.
ON cultures
- 5 ml LB + Cm 25; 37 °C, 225 rpm
- 1C, 2C, 3C, 4C, 5C
BL21 transformation and growth test
Testing BL21 cells with/without CPP expression by transforming and inducing with IPTG.
- 50 μl 0.1 M IPTG
- 40 μl competent cells
- pEX.SOD⋅His
- pEX.nTra10⋅SOD⋅His
- pEX.nTAT⋅SOD⋅His
- pEX.nLMWP⋅SOD⋅His
Mimmi
SOD activity assay
- Kits?
- Biosite/trevigen 6000sek
- Biovision/labinova 3000sek, ca 1week delivery time
- Cayman
- Cellbiolabs
- Merch
- Abcam 3600sek
- Probior
- Sigma-Aldrich 3000sek
- T-cell technology inc.
- Kit from Sigma-Aldrich
- sponsored with a very good offer
 |

|
 |

|
 |

|

|

|
 "
"