Team:Stanford/Digital Sensor
From 2010.igem.org
(→Overview) |
|||
| Line 9: | Line 9: | ||
to calculate the difference in the concentrations of the two input chemicals. One input chemical (chemical A) binds to a promoter and causes the transcription of an mRNA coding for an output protein. The other input chemical (B) binds to a promoter and causes transcription of an sRNA, which is complementary to a target sequence overlapping the ribosome binding site of the mRNA sequence promoted by A. The sRNA then binds to the mRNA, preventing the ribosome from binding and synthesizing the output protein. In the ideal case, no output protein is produced if less A is present than B, and protein begins to be produced as soon as the concentration of A surpasses that of B. To change the threshold ratio detected by the sensor, multiple copies of the genes encoding either the mRNA or the sRNA can be placed downstream of the promoters. | to calculate the difference in the concentrations of the two input chemicals. One input chemical (chemical A) binds to a promoter and causes the transcription of an mRNA coding for an output protein. The other input chemical (B) binds to a promoter and causes transcription of an sRNA, which is complementary to a target sequence overlapping the ribosome binding site of the mRNA sequence promoted by A. The sRNA then binds to the mRNA, preventing the ribosome from binding and synthesizing the output protein. In the ideal case, no output protein is produced if less A is present than B, and protein begins to be produced as soon as the concentration of A surpasses that of B. To change the threshold ratio detected by the sensor, multiple copies of the genes encoding either the mRNA or the sRNA can be placed downstream of the promoters. | ||
| + | |||
| + | Noise abatement: The sRNA sensor and the P/K sensor in digital mode provide at least two antidotes to biological noise: | ||
| + | |||
| + | 1) By sensing only one ratio, we gain a sharp transition between no signal and a lot of signal. This allows us to insulate our output from biological noise in all regimes of the input domain except for the small neighborhood surrounding the sharp transition. | ||
| + | |||
| + | 2) By engineering readout redundancy into the sRNA sensor, we have insulated our sensor from detector-specific readout noise. If there were only one color, we would register the transition from off -> on whenever the signal exceeded the signal/noise ratio of our detector. This means that we would actually read the off/on transition as occurring at different points in the input domain with different detectors! But with two colors, (one transitioning from off -> on, the other from on -> off), we can set the detection threshold far above our detector's S/N threshold, and average the transition point in the two channels to achieve a detector-independent measurement of the specified ratio. | ||
Read about our [[Team:Stanford/Analog Sensor|other sensor]] or return to our [[Team:Stanford/Research| project]] page. | Read about our [[Team:Stanford/Analog Sensor|other sensor]] or return to our [[Team:Stanford/Research| project]] page. | ||
Revision as of 07:01, 27 October 2010

| Home | Project | Applications | Modeling | Parts | Team | Notebook |
The sRNA Sensor
Overview
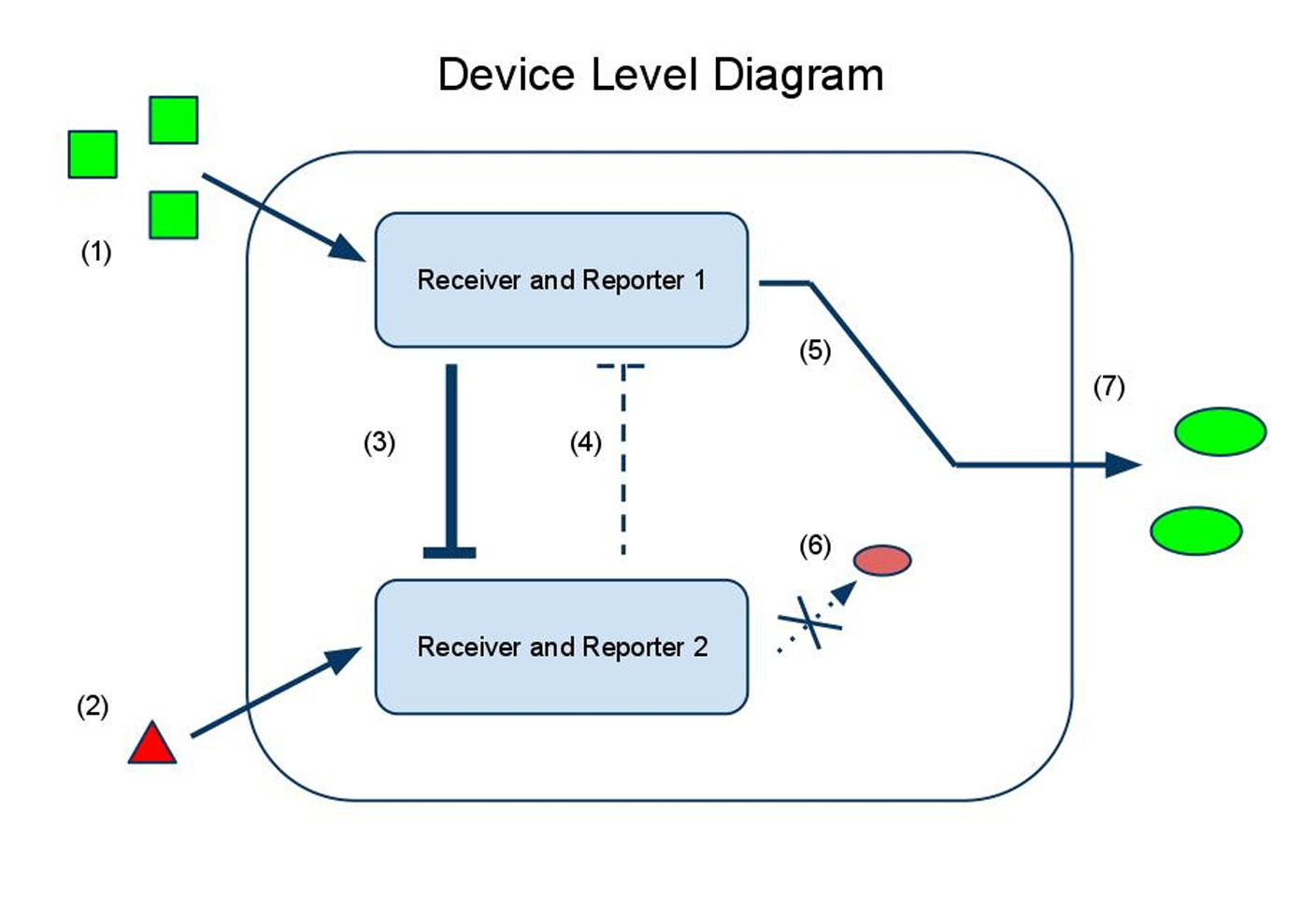
Our first sensor design is tuned to a single ratio of input chemicals and tells the user if the input chemicals are present in relative concentrations below, at, or above that ratio.
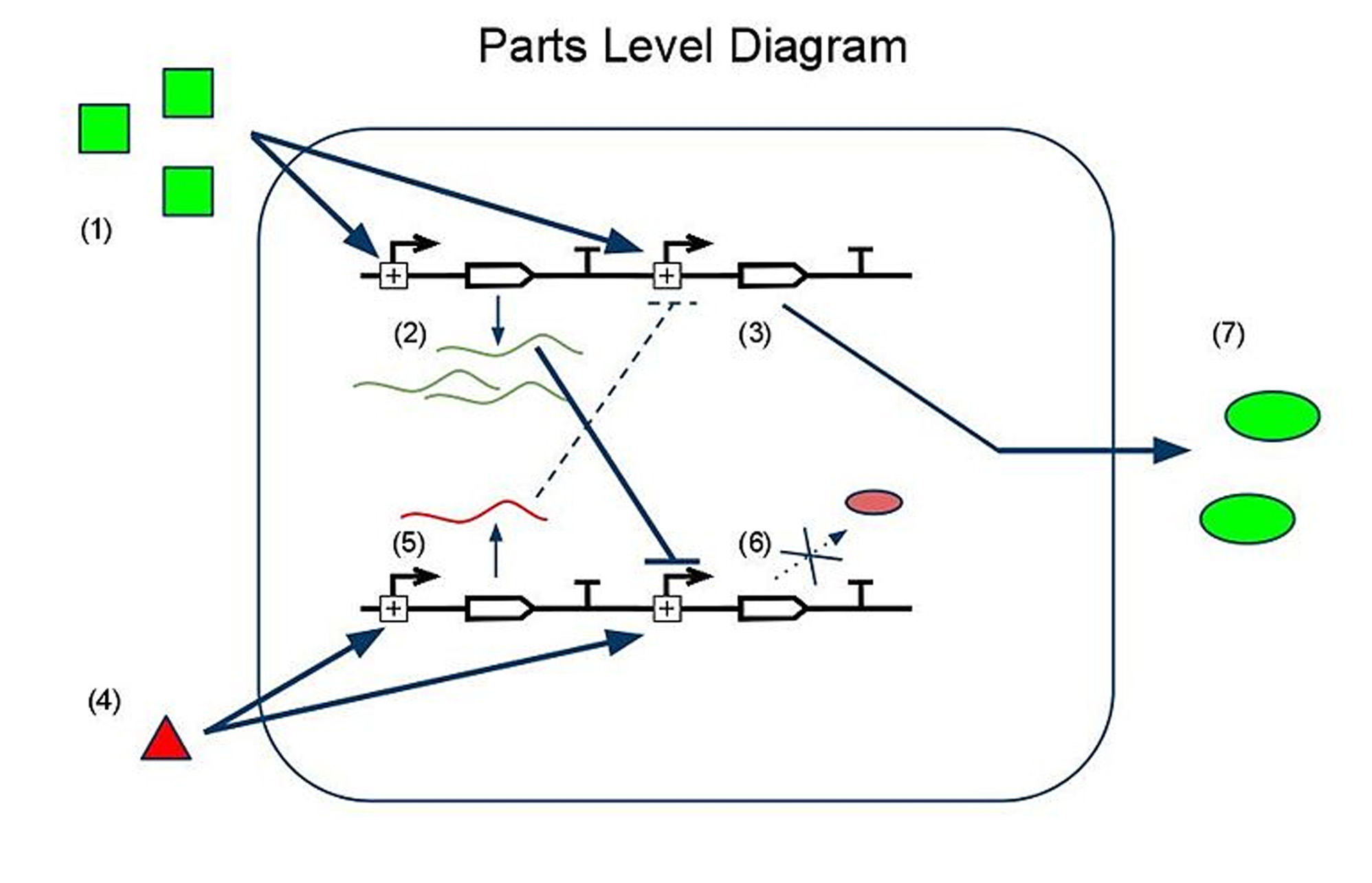
to calculate the difference in the concentrations of the two input chemicals. One input chemical (chemical A) binds to a promoter and causes the transcription of an mRNA coding for an output protein. The other input chemical (B) binds to a promoter and causes transcription of an sRNA, which is complementary to a target sequence overlapping the ribosome binding site of the mRNA sequence promoted by A. The sRNA then binds to the mRNA, preventing the ribosome from binding and synthesizing the output protein. In the ideal case, no output protein is produced if less A is present than B, and protein begins to be produced as soon as the concentration of A surpasses that of B. To change the threshold ratio detected by the sensor, multiple copies of the genes encoding either the mRNA or the sRNA can be placed downstream of the promoters.
Noise abatement: The sRNA sensor and the P/K sensor in digital mode provide at least two antidotes to biological noise:
1) By sensing only one ratio, we gain a sharp transition between no signal and a lot of signal. This allows us to insulate our output from biological noise in all regimes of the input domain except for the small neighborhood surrounding the sharp transition.
2) By engineering readout redundancy into the sRNA sensor, we have insulated our sensor from detector-specific readout noise. If there were only one color, we would register the transition from off -> on whenever the signal exceeded the signal/noise ratio of our detector. This means that we would actually read the off/on transition as occurring at different points in the input domain with different detectors! But with two colors, (one transitioning from off -> on, the other from on -> off), we can set the detection threshold far above our detector's S/N threshold, and average the transition point in the two channels to achieve a detector-independent measurement of the specified ratio.
Read about our other sensor or return to our project page.
Diagrams
 "
"