Team:Slovenia/PROJECT/proof/studies/spr
From 2010.igem.org
Contents |
Introduction
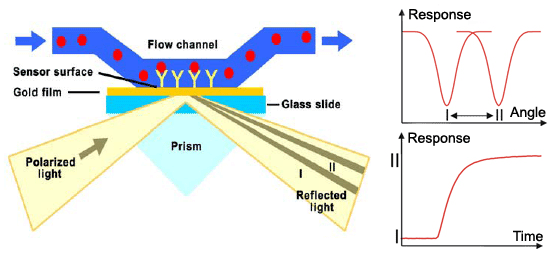
Surface plasmon resonance (SPR) is a method used for detailed qualitative and quantitave analysis of molecular interactions, in our case binding of zinc fingers to DNA. Method uses surface plasmons (also called surface plasmon polaritons) – surface electromagnetic waves that propagate parallel to the metal/dielectric interface, most commonly gold and silver. Waves, due to direction and location on the boundary between metal and external medium, are very sensitive to any changes occurring on the boundary. Electron or light beam is used to excite surface plasmons to resonate properly. System detects changes in the refractive index of the surface layer of a solution. SPR is observed as a sharp dip in reflected intensity at an angle which depends on the refractive index of the medium on the non-iluminated side of the surface. The angle shifts when biomolecules bind to the surface and change the refractive index of the surface layer.
The sensorgram is a plot of the SPR angle against time, and displays the real-time progress of the interaction at the sensor surface. Main advantages of this method include: no labeling needed, real-time measurements, small amounts of the samples required, reusable sensor chips, etc. The method can measure the interactions between proteins, protein-DNA, protein-membrane, receptor-low molecular analytes,… The information obtained from the sensorgrams is:
- Specificity - how specific is the binding between two molecules
- Concentration - how much of a given molecule is bound and/or active
- Binding affinity of the analyte to ligand
- Kinetics - the rate of association and dissociation
Our SPR system
Our main aim was to show that the selected synthetic zinc fingers bind to their respective DNA targets. We used Biacore T100 (GE Healthcare) and avidin-coated sensor chip which served as a platform for binding of a biotinylated adaptor oligonucleotide. This oligo was designed in a way that it formed a short nucleotide overlap with DNA program sequence thereby enabling us to bind it reversibly each time zinc finger binding events were tested. This fact was of high importance since when testing single binding events, we wanted to have all binding sites within immobilized DNA program unoccupied. When binding more than one zinc finger sequentially we wanted the opposite - to have all binding sites for a particular zinc finger occupied - therefore being sure the second zinc finger injected would bound adjacent DNA target element on a DNA program preoccupied by the first one. Since we proved that zinc finger fusions exhibit successful binding, this was also a good indication that zinc finger enzyme fusions will bind their respective DNA target sites in vivo.
Results
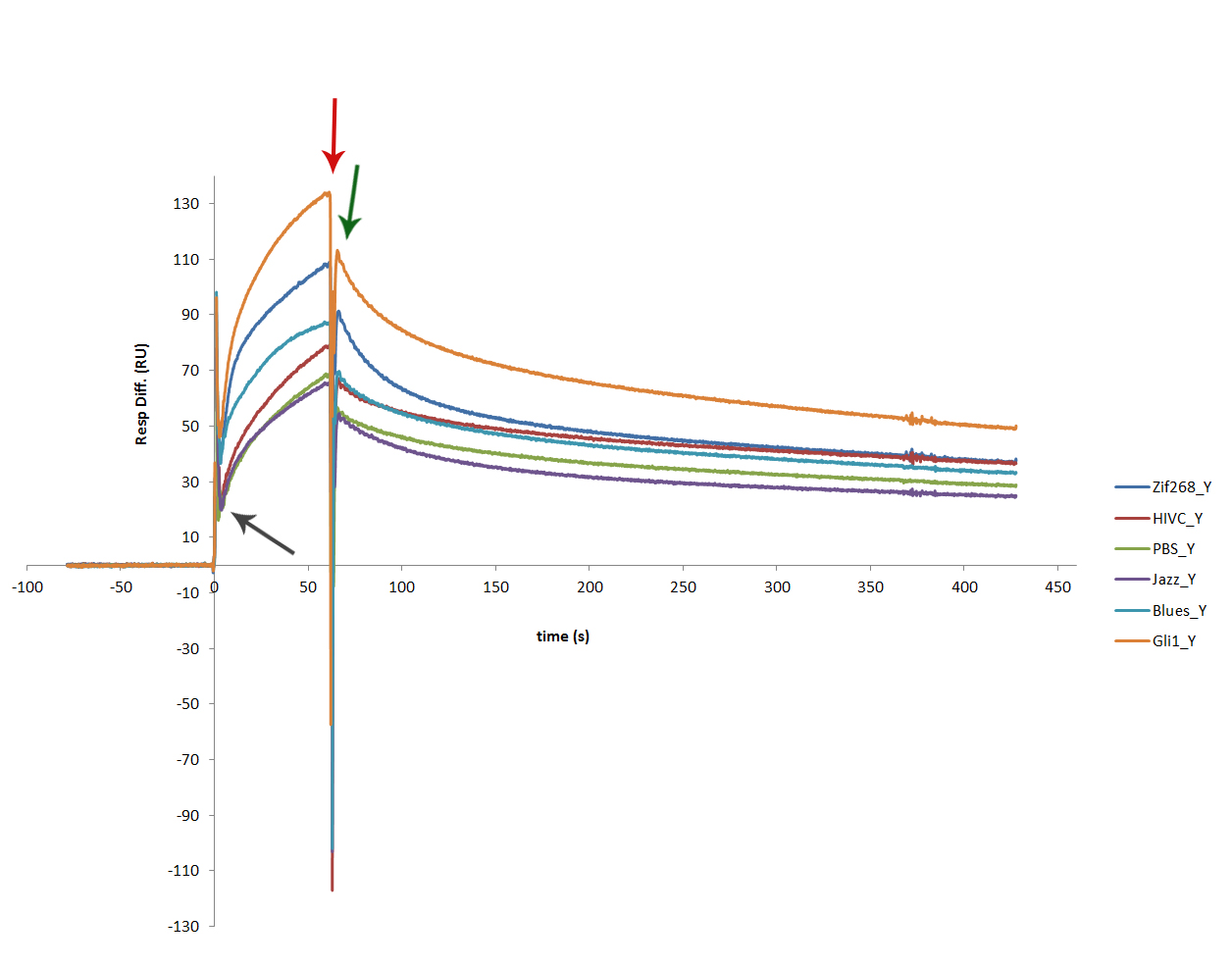
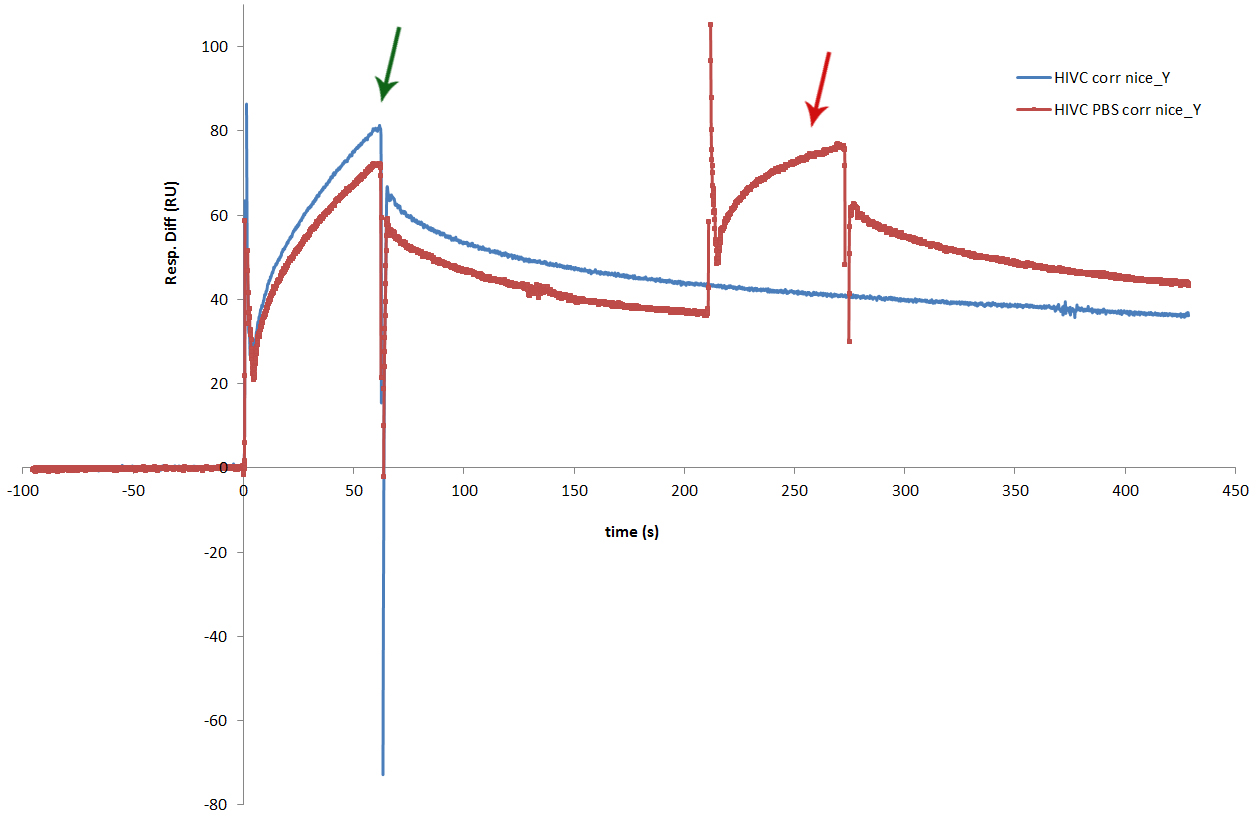
The proteins dissolved from the inclusion bodies were tested with SPR. SPR experiment showed that each of zinc fingers used in our experiments, successfully binds to its binding sequence. Sensorgram obtained from the experiment shows similar binding results for all 6 zinc fingers [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323069 cCFP_link_HIVC], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323058 Gli1_link_nCFP], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323001 cYFP_link_Zif268], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323015 PBSII_link_nYFP], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323070 Blues_link_nCF], [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323004 cCFP_link_Jazz].
 "
"