Team:Slovenia/PROJECT/proof/program
From 2010.igem.org
(New page: {{Team:Slovenia/templatetop}} {{Team:Slovenia/templateproject}} <html> <style> #vsebina_mid{ height: 2000px; } #subgumb3{display:none}#subgumb4{display:none}#subgumb5{display:none}#subgumb...) |
|||
| (21 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<style> | <style> | ||
#vsebina_mid{ | #vsebina_mid{ | ||
| - | height: | + | height: 3050px; |
| + | } | ||
| + | |||
| + | #lgumb2{ | ||
| + | background-image:url("https://static.igem.org/mediawiki/2010/3/3e/SLOproofover.png"); | ||
| + | } | ||
| + | #subgumb1{ | ||
| + | |||
| + | background-image:url("https://static.igem.org/mediawiki/2010/d/d7/SLODNAprogramover.png"); | ||
} | } | ||
#subgumb3{display:none}#subgumb4{display:none}#subgumb5{display:none}#subgumb6{display:none}#subgumb7{display:none}#subgumb8{display:none}#subgumb9{display:none}#subgumb10{display:none} | #subgumb3{display:none}#subgumb4{display:none}#subgumb5{display:none}#subgumb6{display:none}#subgumb7{display:none}#subgumb8{display:none}#subgumb9{display:none}#subgumb10{display:none} | ||
</style> | </style> | ||
<div id="overhead"> | <div id="overhead"> | ||
| - | <div id="naslov"> | + | <div id="naslov">DNA program</div> |
</div> | </div> | ||
<div id="besedilo"> | <div id="besedilo"> | ||
</html> | </html> | ||
| + | |||
<!-- OD TU NAPREJ PIŠI BESEDILO --> | <!-- OD TU NAPREJ PIŠI BESEDILO --> | ||
| - | < | + | __TOC__ |
| - | + | ||
| - | < | + | <h2>What is a DNA program?</h2> |
| - | + | ||
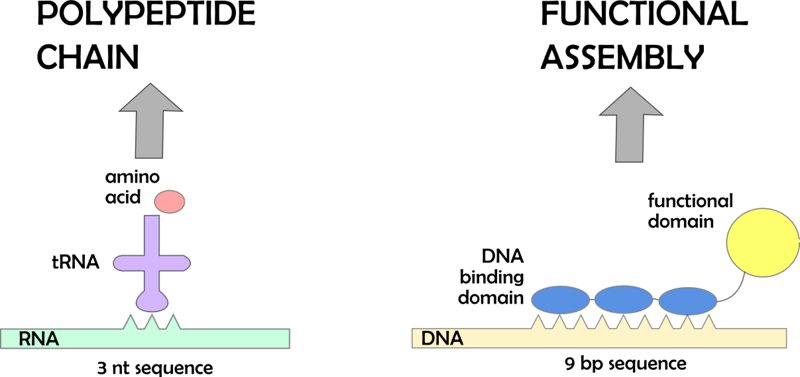
| - | + | DNA program is a noncoding DNA sequence consisting of specific binding sites for DNA binding domains. These DNA binding domains can be linked to functional domains (e.g. enzymes). This is analogous to the recognition of RNA codons by anticodons on aminoacyl tRNA, where the aminoacyl tRNA corresponds to the DNA binding domain with functional protein domain. When those functional domains assemble on a DNA program they are brought closer together in a defined order. By changing the places of binding sites on a DNA program it is possible to change the sequence of events (e.g. course of the reaction in case of the enzymes). <br> | |
| - | + | ||
| - | + | ||
| - | + | [[Image:Slo tRNA gor.png|thumb|center|550px|'''Figure 1:''' Analogy between central biological dogma and our idea of coding beyond triplets is preseneted]] | |
| - | + | ||
| - | + | ||
| - | + | <h2>Selection and importance of spacer sites</h2> | |
| - | We | + | |
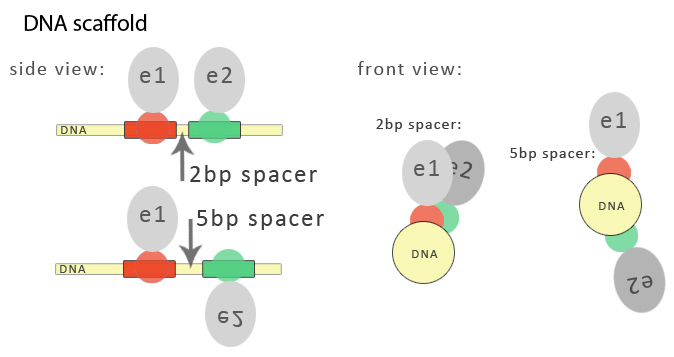
| - | < | + | Binding sites for three-fingered zinc fingers span 9 nucleotides but can be extended to 18 base pair recognition motifs for longer zinc fingers, spanning from one to two DNA duplex helical turns, respectively. Binding sites for DNA-binding proteins are separated by spacers, which are nucleotides that are not occupied by DNA-binding proteins. The length of the spacer sequence is not coincidental. The selection follows three dimensional structure of a DNA molecule. One turn of DNA helix is 10,5 base pairs long, which roughly overlaps with the length of a DNA molecule encircled by one zinc finger domain recognising and binding to 9 base pairs. In order to have functional units on the same side of a DNA molecule serving as a DNA program, it is of high importance to select the right spacer length. This is the case when having split functional units attached to DNA binding domains as well as when biosynthetic enzymes are selected for functional domains. Double helix of DNA defines on which side of the helix the functional domain will be attached, which is defined by the length of the spacer between DNA domain binding sites: spacer of 1 or 2 nucleotides positions them very close, while the spacer of five nucleotides positions the neighboring two functional domains to the opposite sides of the helix.<br> |
| - | + | ||
| - | < | + | [[Image:slo_dna_scafold2.jpg|thumb|center|700px|'''Figure 2:''' Role of the spacer length between binding sites in a DNA program.]] |
| - | + | ||
| - | + | We employed an in-house computer software in order to predict appropriate spacer sites, which selects the base pairs in such a way that overlapping of recognition motifs wouldn't occur and disturb the sequential binding of selected synthetic zinc fingers. Two base pair spacer sequence was selected by default based on the literature since it has been shown that it leads to efficient split GFP reassembly. Increased length of a spacer would be particularly useful for the assembly of large protein functional domains that exceed 3.5 nm, which is the pitch of B-type DNA duplex.<br><br> | |
| - | + | ||
| - | < | + | <h2>Variability of DNA program</h2> |
| - | The | + | |
| - | < | + | Another quality of the idea of DNA program is it's variability. If we take only the most characterized DNA binding domains that bind 9 base pair motifs, they can in theory form 262.144 unique combinations. This is over 4000-fold increase over 64 possibilities within a DNA triplet code. Furthermore, DNA binding domains have been characterised that bind to 18 base pairs which increases the possibilities even further. The main advantage of our approach is that the sequence of DNA binding motifs are basically not constrained, we can select a DNA sequence for which we have the well characterized DNA binding protein available. <br><br> |
| - | + | ||
| - | < | + | <h2>Applications of DNA programs and beyond</h2> |
| - | + | ||
| - | [[Image: | + | The most promising direction of the application of DNA programs probably lies in biosynthetic pathways. The approach can be applied to many other biosynthetic pathways where enhanced production and/or intermediate substrate channelling is desired to avoid unwanted metabolite flows. A possibility of implementing simple information processing circuits such as DNA based logical gates using split/FRET system can also be envisaged but was not further investigated during the project.<br><br> |
| - | + | ||
| - | + | <h2>Cloning strategy to increase DNA program copy number</h2> | |
| - | + | ||
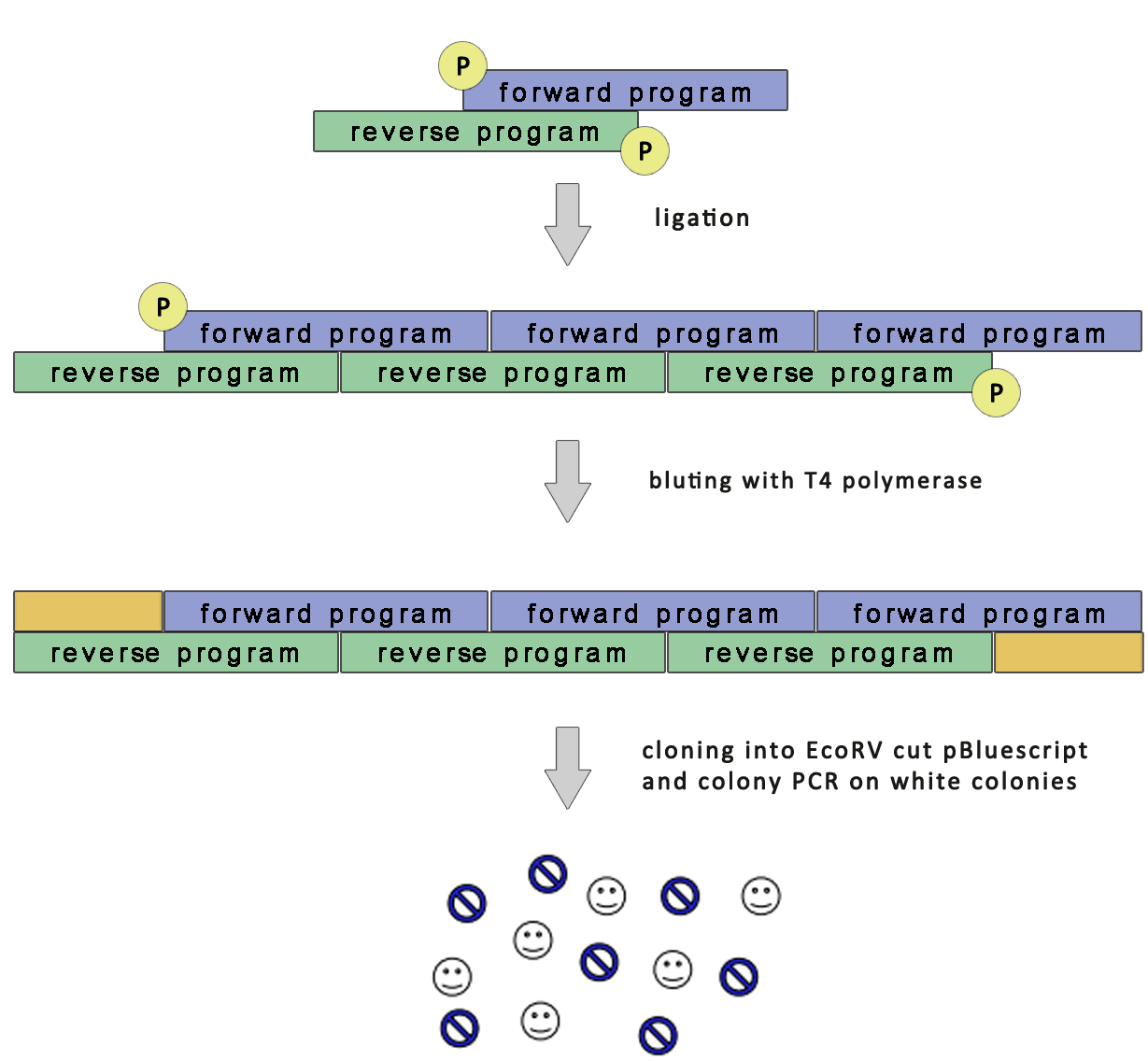
| - | + | For the increased production of biosynthetic products multiple copies of DNA program should be beneficial. We employed cloning of multiple copies of a DNA program simultaneously. This was achieved by ordering overlapping 5' phosphorylated DNA program primers. When annealed they were ligated, blunt-ended with T4 polymerase and cloned into the vector. A scheme of the cloning strategy is shown below:<br> | |
| + | |||
| + | [[Image:slo_program.png|thumb|center|700px|'''Figure 3:''' Cloning strategy to increase DNA program copy number.]] | ||
<!--STOP BESEDILO--> | <!--STOP BESEDILO--> | ||
Latest revision as of 00:07, 28 October 2010
Contents |
What is a DNA program?
DNA program is a noncoding DNA sequence consisting of specific binding sites for DNA binding domains. These DNA binding domains can be linked to functional domains (e.g. enzymes). This is analogous to the recognition of RNA codons by anticodons on aminoacyl tRNA, where the aminoacyl tRNA corresponds to the DNA binding domain with functional protein domain. When those functional domains assemble on a DNA program they are brought closer together in a defined order. By changing the places of binding sites on a DNA program it is possible to change the sequence of events (e.g. course of the reaction in case of the enzymes).
Selection and importance of spacer sites
Binding sites for three-fingered zinc fingers span 9 nucleotides but can be extended to 18 base pair recognition motifs for longer zinc fingers, spanning from one to two DNA duplex helical turns, respectively. Binding sites for DNA-binding proteins are separated by spacers, which are nucleotides that are not occupied by DNA-binding proteins. The length of the spacer sequence is not coincidental. The selection follows three dimensional structure of a DNA molecule. One turn of DNA helix is 10,5 base pairs long, which roughly overlaps with the length of a DNA molecule encircled by one zinc finger domain recognising and binding to 9 base pairs. In order to have functional units on the same side of a DNA molecule serving as a DNA program, it is of high importance to select the right spacer length. This is the case when having split functional units attached to DNA binding domains as well as when biosynthetic enzymes are selected for functional domains. Double helix of DNA defines on which side of the helix the functional domain will be attached, which is defined by the length of the spacer between DNA domain binding sites: spacer of 1 or 2 nucleotides positions them very close, while the spacer of five nucleotides positions the neighboring two functional domains to the opposite sides of the helix.
We employed an in-house computer software in order to predict appropriate spacer sites, which selects the base pairs in such a way that overlapping of recognition motifs wouldn't occur and disturb the sequential binding of selected synthetic zinc fingers. Two base pair spacer sequence was selected by default based on the literature since it has been shown that it leads to efficient split GFP reassembly. Increased length of a spacer would be particularly useful for the assembly of large protein functional domains that exceed 3.5 nm, which is the pitch of B-type DNA duplex.
Variability of DNA program
Another quality of the idea of DNA program is it's variability. If we take only the most characterized DNA binding domains that bind 9 base pair motifs, they can in theory form 262.144 unique combinations. This is over 4000-fold increase over 64 possibilities within a DNA triplet code. Furthermore, DNA binding domains have been characterised that bind to 18 base pairs which increases the possibilities even further. The main advantage of our approach is that the sequence of DNA binding motifs are basically not constrained, we can select a DNA sequence for which we have the well characterized DNA binding protein available.
Applications of DNA programs and beyond
The most promising direction of the application of DNA programs probably lies in biosynthetic pathways. The approach can be applied to many other biosynthetic pathways where enhanced production and/or intermediate substrate channelling is desired to avoid unwanted metabolite flows. A possibility of implementing simple information processing circuits such as DNA based logical gates using split/FRET system can also be envisaged but was not further investigated during the project.
Cloning strategy to increase DNA program copy number
For the increased production of biosynthetic products multiple copies of DNA program should be beneficial. We employed cloning of multiple copies of a DNA program simultaneously. This was achieved by ordering overlapping 5' phosphorylated DNA program primers. When annealed they were ligated, blunt-ended with T4 polymerase and cloned into the vector. A scheme of the cloning strategy is shown below:
 "
"