Team:Slovenia/PROJECT/proof/popfret/split
From 2010.igem.org
| Line 4: | Line 4: | ||
<style> | <style> | ||
#vsebina_mid{ | #vsebina_mid{ | ||
| - | height: | + | height: 5000px; |
} | } | ||
#lgumb2{ | #lgumb2{ | ||
| Line 69: | Line 69: | ||
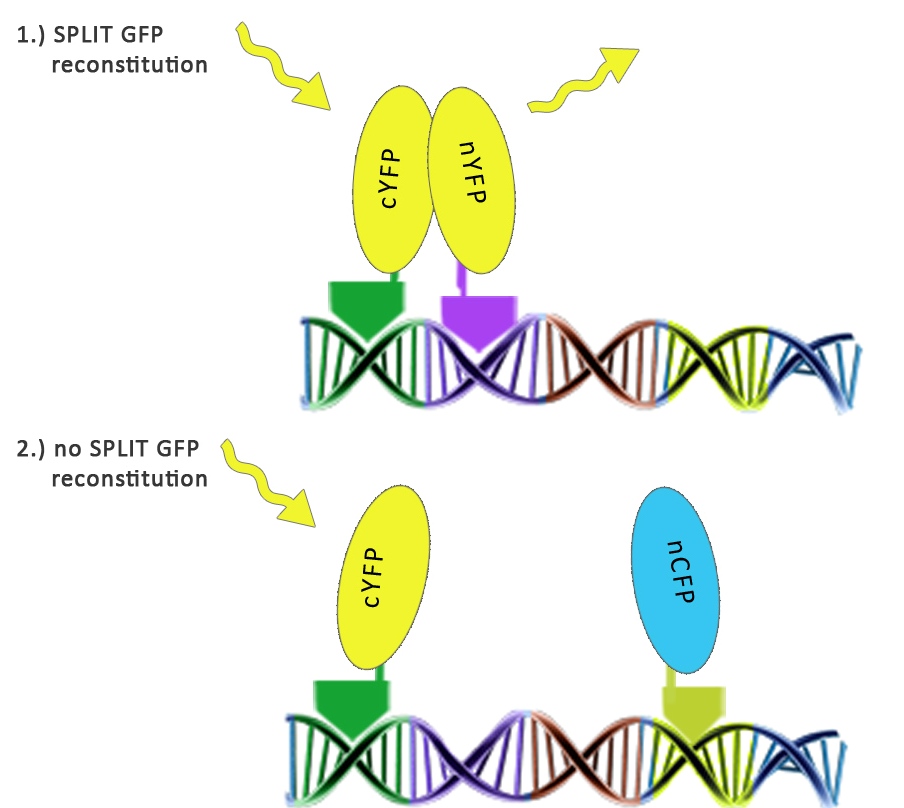
We used split fluorescent proteins fused with zinc fingers to demonstrate the need of a DNA program for fluorescent protein reassembly. To our knowledge, reconstituition of split GFPs using DNA program has never been demonstrated <em>in vivo</em>, but only <em>in vitro </em>(Segal et al., 2005). In addition we wanted to demonstrate that the distance between zinc finger binding sites plays an important role in split GFP reconstitution. Moreover, we tested split GFP (mCerulean and mCitrine) reassembly<em> in vitro</em> using cell lysates. We selected mCitrine, a yellow variant of GFP for the initial experiments of reconstitution of split GFPs. mCitrine is brighter than enhanced yellow GFP and less sensitive to lower pH than EYFP or mVenus. | We used split fluorescent proteins fused with zinc fingers to demonstrate the need of a DNA program for fluorescent protein reassembly. To our knowledge, reconstituition of split GFPs using DNA program has never been demonstrated <em>in vivo</em>, but only <em>in vitro </em>(Segal et al., 2005). In addition we wanted to demonstrate that the distance between zinc finger binding sites plays an important role in split GFP reconstitution. Moreover, we tested split GFP (mCerulean and mCitrine) reassembly<em> in vitro</em> using cell lysates. We selected mCitrine, a yellow variant of GFP for the initial experiments of reconstitution of split GFPs. mCitrine is brighter than enhanced yellow GFP and less sensitive to lower pH than EYFP or mVenus. | ||
| - | [[Image:slo_split_prava.jpg|thumbs|center| | + | [[Image:slo_split_prava.jpg|thumbs|center|600px]] |
<h2>Results</h2> | <h2>Results</h2> | ||
| - | [[Image:split_yfp_brez.png|thumbs|center| | + | [[Image:split_yfp_brez.png|thumbs|center|500px]] |
| - | [[Image:split_yfp_z.png|thumbs|center| | + | [[Image:split_yfp_z.png|thumbs|center|500px]] |
| + | |||
| + | Split YFP reconstitution occurs only in the presence of the appropriate DNA program. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323005 PBSII_link_nYFP] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268], 25ng each, were cotransfected into HEK293 cells in the (a) absence and (b) presence of 500 ng of DNA program sequence in [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 pSB1C3 backbone]. The HEK293 cells were excited with 514-nm Ar-laser and emission was detected between 520 and 580 nm. | ||
| + | |||
| - | |||
Revision as of 16:14, 27 October 2010
Contents |
Introduction
We used split fluorescent proteins fused with zinc fingers to demonstrate the need of a DNA program for fluorescent protein reassembly. To our knowledge, reconstituition of split GFPs using DNA program has never been demonstrated in vivo, but only in vitro (Segal et al., 2005). In addition we wanted to demonstrate that the distance between zinc finger binding sites plays an important role in split GFP reconstitution. Moreover, we tested split GFP (mCerulean and mCitrine) reassembly in vitro using cell lysates. We selected mCitrine, a yellow variant of GFP for the initial experiments of reconstitution of split GFPs. mCitrine is brighter than enhanced yellow GFP and less sensitive to lower pH than EYFP or mVenus.
Results
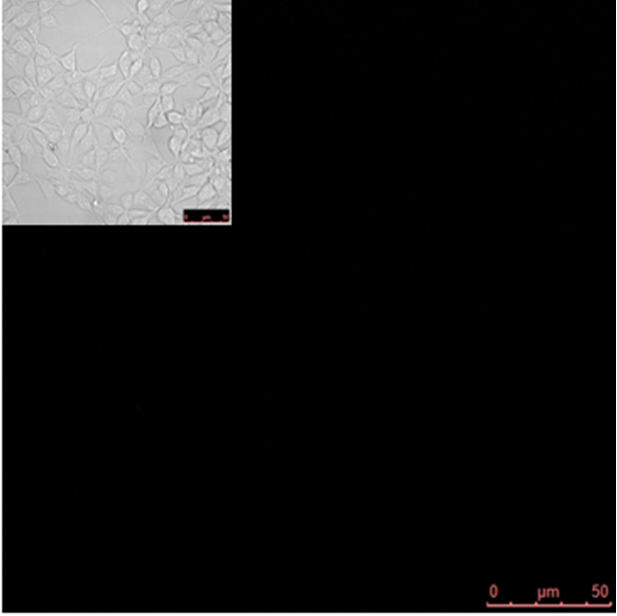
Split YFP reconstitution occurs only in the presence of the appropriate DNA program. [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323005 PBSII_link_nYFP] and [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323080 cYFP_link_Zif268], 25ng each, were cotransfected into HEK293 cells in the (a) absence and (b) presence of 500 ng of DNA program sequence in [http://partsregistry.org/wiki/index.php?title=Part:BBa_K323039 pSB1C3 backbone]. The HEK293 cells were excited with 514-nm Ar-laser and emission was detected between 520 and 580 nm.
 "
"