Team:Slovenia/PROJECT/biosynthesis/carotenoids
From 2010.igem.org
| (21 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<style> | <style> | ||
#vsebina_mid{ | #vsebina_mid{ | ||
| - | height: | + | height: 8150px; |
} | } | ||
#lgumb3{ | #lgumb3{ | ||
| Line 21: | Line 21: | ||
<!-- OD TU NAPREJ PIŠI BESEDILO --> | <!-- OD TU NAPREJ PIŠI BESEDILO --> | ||
| - | + | __TOC__ | |
| - | + | <div id="okvircek" style="background-color:#CECDCD;border:1px solid #AEABAE;margin-top:9px;padding:11px"> | |
| + | <h3>State of the art</h3> | ||
<ul> | <ul> | ||
| Line 28: | Line 29: | ||
</ul> | </ul> | ||
| - | + | <h3>Aims</h3> | |
<ul> | <ul> | ||
| - | <li> | + | <li>Add additional genes enzymes and achieve biosynthesis of carotenoids zeaxanthin and canthaxanthin</li> |
| - | <li> | + | <li>Prepare DNA-guided biosynthetic pathways for production of zeaxanthin and canthaxanthin in ''E.coli''</li> |
</ul> | </ul> | ||
| - | + | <h3>Achievements</h3> | |
<ul> | <ul> | ||
| - | <li> | + | <li>We demonstrated production of zeaxanthin in <em>E.coli</em> using parts from the Registry</li> |
| - | <li> | + | <li>We added an additional carotenoid biosynthetic pathway enzyme crtO and demonstrated formation of a carotenoid that was probably echinenone</li> |
| - | <li> | + | <li>We constructed DNA-guided biosynthetic pathway constructs but did not have time to test it experimentally as the cells with three plasmids grow extremely slow</li> |
</ul> | </ul> | ||
| + | </div> | ||
| + | <h2>Background</h2> | ||
| - | + | Carotenoids form a large family of colored compounds, naturally occuring in plants and bacteria. Besides being vitamin A precursors, many carotenoids have a potent antioxidant activity and are therefore widely used as food supplements, food colorants in cosmetics and as animal feed. According to BCC Research total global market for carotenoids was worth $766 million in 2007 and is expected to increase to $909 million till 2015. The largest share goes to beta carotene, with asthaxanthin closely following and canthaxanthin in the third place. | |
| - | + | ||
| - | Carotenoids form a large family of colored compounds, naturally occuring in plants and bacteria. Besides being vitamin A precursors , many carotenoids have a potent antioxidant activity and are therefore widely used as food supplements, food colorants in cosmetics and as animal feed. According to BCC Research total global market for carotenoids was worth $766 million in 2007 and is expected to increase to $909 million till 2015. The largest share goes to beta carotene, with asthaxanthin closely following and canthaxanthin in the third place. | + | |
Due to difficulties in obtaining some carotenoids from natural sources on a large scale, they are produced by chemical synthesis. But especially in food industry they are generally preferred from natural sources over synthetic compounds. Also due to the cost and sustainability, there is a large interest to use microbial hosts for carotenoid biosynthesis. | Due to difficulties in obtaining some carotenoids from natural sources on a large scale, they are produced by chemical synthesis. But especially in food industry they are generally preferred from natural sources over synthetic compounds. Also due to the cost and sustainability, there is a large interest to use microbial hosts for carotenoid biosynthesis. | ||
| - | + | <h2>Biosynthetic pathway</h2> | |
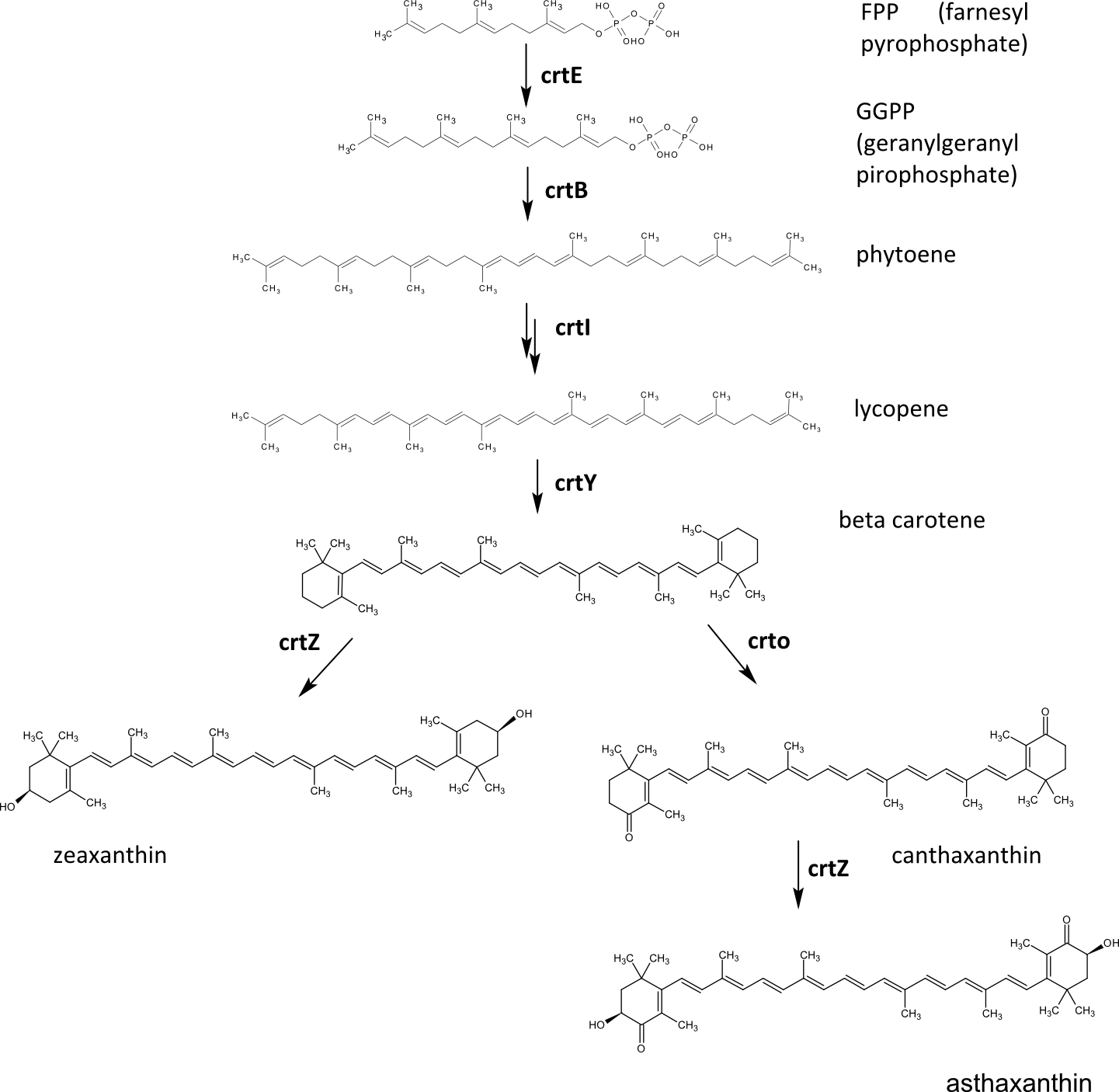
Many carotenoids have been produced in <em>E.coli</em> using reconstitution of a plant biosynthetic pathway in bacteria. The precursor for carotenoids is farnesyl pyrophoshpate. The first three enzymes in the biosynthetic pathway produce lycopene, a red pigment found in tomatoes. The next enzyme in the pathway, lycopene beta-cyclase (crtY), can transform it to beta carotene, which is a vitamin A precursor. This segment of the carotenoid biosynthetic pathway was already available in the Registry and iGEM teams from previous years (Edinburgh 07, Edinburgh 08, Guelph 08, Cambridge09) demonstrated production of beta-carotene and lycopene. | Many carotenoids have been produced in <em>E.coli</em> using reconstitution of a plant biosynthetic pathway in bacteria. The precursor for carotenoids is farnesyl pyrophoshpate. The first three enzymes in the biosynthetic pathway produce lycopene, a red pigment found in tomatoes. The next enzyme in the pathway, lycopene beta-cyclase (crtY), can transform it to beta carotene, which is a vitamin A precursor. This segment of the carotenoid biosynthetic pathway was already available in the Registry and iGEM teams from previous years (Edinburgh 07, Edinburgh 08, Guelph 08, Cambridge09) demonstrated production of beta-carotene and lycopene. | ||
| Line 57: | Line 58: | ||
Another biosynthetic pathway of beta carotene modification is oxidation by the enzyme beta ketolase (crtO), which results in another industrially interesting compound - canthaxanthin. This enzyme, however, preferrably oxidizes only one ring of the beta carotene, which leads to formation of carotenoid called echinenone, but when the gene is overexpressed, canthaxanthin can be produced as well. Canthaxanthin can also be further modified by beta hydroxylase, which gives red pigment asthaxanthin as a product. | Another biosynthetic pathway of beta carotene modification is oxidation by the enzyme beta ketolase (crtO), which results in another industrially interesting compound - canthaxanthin. This enzyme, however, preferrably oxidizes only one ring of the beta carotene, which leads to formation of carotenoid called echinenone, but when the gene is overexpressed, canthaxanthin can be produced as well. Canthaxanthin can also be further modified by beta hydroxylase, which gives red pigment asthaxanthin as a product. | ||
| - | [[Image:SLO_shema_karotenoidi_brez_programa.png|thumb|center|700px|'''Scheme of the carotenoid biosynthesis pathways with enzymes, used in our project. | + | [[Image:SLO_shema_karotenoidi_brez_programa.png|thumb|center|700px|'''Figure 1:''' Scheme of the carotenoid biosynthesis pathways with enzymes, used in our project.]] |
<h2>Aims</h2> | <h2>Aims</h2> | ||
| - | + | ||
| - | + | We wanted to improve the velocity, yield and specificity of carotenoid biosynthesis in ''E. coli'' using the DNA-guided biosynthetic enzyme ordered assembly. We constructed genes coding for chimeric enzyme - zinc finger fusion proteins (see scheme below) and designed two DNA programs, which specifically assemble carotenoid biosynthetic enzymes of each pathway. A sequence of crtE, crtB, crtI, crtY, crtO and crtZ enzymes should produce asthaxanthin, while crtE, crtB, crtI, crtY and crtZ should result in the production of zeaxanthin. (see scheme below). | |
| - | + | ||
| - | + | [[Image:shema_karotenoidi.png|thumb|center|700px|'''Figure 2:''' Scheme of desired outcome of the carotenoid biosynthesis with two distinct DNA programs.]] | |
| - | + | ||
| - | + | ||
| - | + | One of the reasons why carotenoid biosynthesis pathway represented a challenge for us was putative membrane localisation of at least a few enzymes of the pathway. The literature does not provide the answer to this question. Membrane localization of some enzymes would mean that some enzymes are in a way already compartmentalized in the cell, but we nevertheless expected some improvement using DNA guided assembly. Another question regarding the membrane localization of the enzymes was whether they will retain their activity if fused to zinc finger domain, as additional domain at the N-terminus could disturb their membrane association. | |
| - | + | ||
| - | <h2 | + | <h2>Cloning strategy</h2> |
| - | + | ||
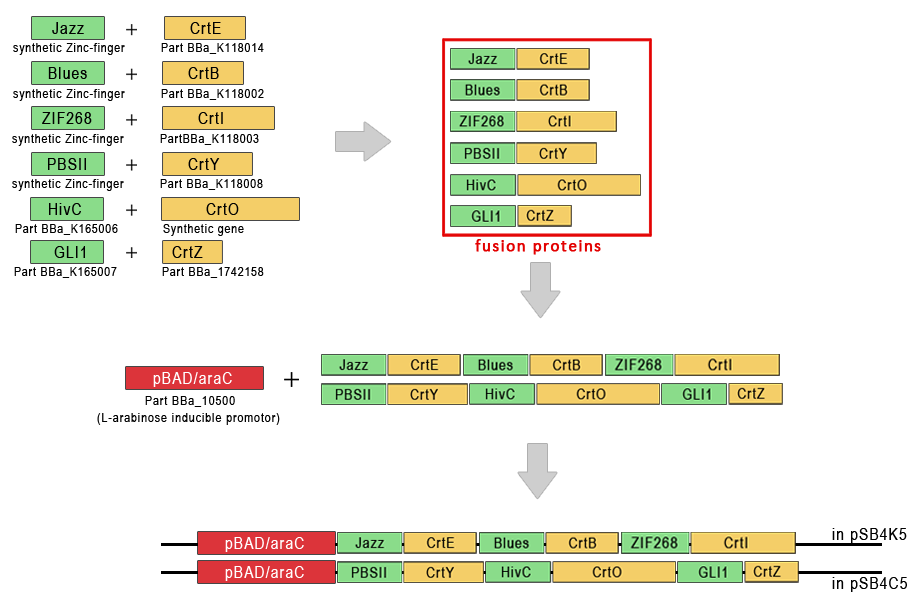
| - | + | As the sum of lengths of genes coding for chimeric enzymes for carotenoid biosynthesis is almost 9 kbp, we decided to clone them into two separate plasmids with different antibiotic resistance. We cloned the first three genes (Jazz::crtE, Blues::crtB, Zif268::crtI) into a vector with kanamycin resistance (pSB4K5) and the last three (PSBII::crtY, ZnfHivC::crtO and crtZ::Gli1) into a vector with chloramphenicol resistance (pSB4C5). | |
| - | + | ||
| - | + | To faciliate the cloning process we designed our chimeric enzymes with zinc finger domain N-terminal to the enzymes. The genes for enzymes from The Registry include both start and stop codon, which meant we could directly fuse them to C-terminus of our synthetic zinc finger domains which do not have stop codon. | |
| - | + | ||
| - | + | We chose a linker olygopeptide of 10 amino acids, consisting of Gly and Ser residues to connect zinc fingers and enzymes, because it enables the flexibility and has also been used for similar purposes before. DNA program encoding the sequence of chimeric enzymes was introduced into bacteria on the third plasmid. | |
| - | + | ||
| - | <h2 | + | [[Image:SLOkarotenoidi_shema.png|thumb|center|700px|'''Figure 3:''' Scheme of the cloning strategy for carotenoid biosynthesis based on DNA-guided pathway assembly.]] |
| - | + | ||
| - | < | + | <h2>Results</h2> |
| - | + | ||
| - | + | <h3>Biosynthesis of zeaxanthin</h3> | |
| - | + | ||
| - | + | We demonstrated that enzyme crtZ can produce zeaxanthin from beta carotene by inserting the gene, coding for this enzyme, behind the beta carotene-operon, submitted to Registry of parts by teams from previous years. This extends the verified carotenoid biosynthetic pathway in ''E.coli'' available in the Registry for an additional step. | |
| - | + | ||
| - | + | [[Image:SLO_HPLC_zeaxanthin.jpg|thumb|center|700px|'''Figure 4:''' HPLC analysis of extracts from bacteria comprising the zeaxanthin biosynthetic pathway (R0011 crtEBIYZ) confirms production of zeaxanthin.]] | |
| - | + | ||
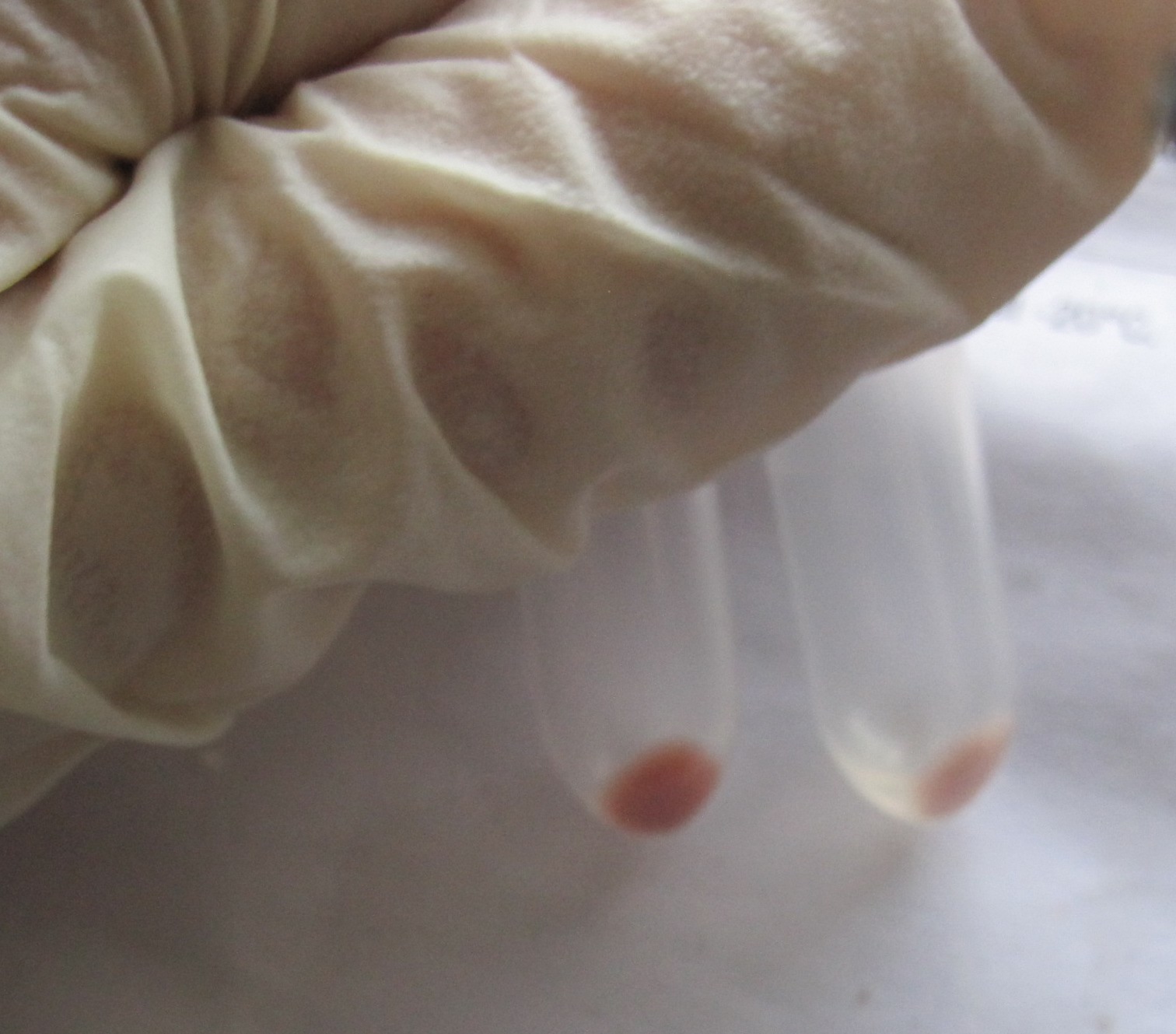
| - | + | [[Image:SLO_pelet_zeaxanthin.JPG|thumb|center|350px|'''Figure 5:''' Cell pellets of bacterial culture producing zeaxanthin (R0011 crtEBIYZ)]] | |
| - | + | ||
| - | < | + | <h3>Biosynthesis of canthaxanthin</h3> |
| - | + | ||
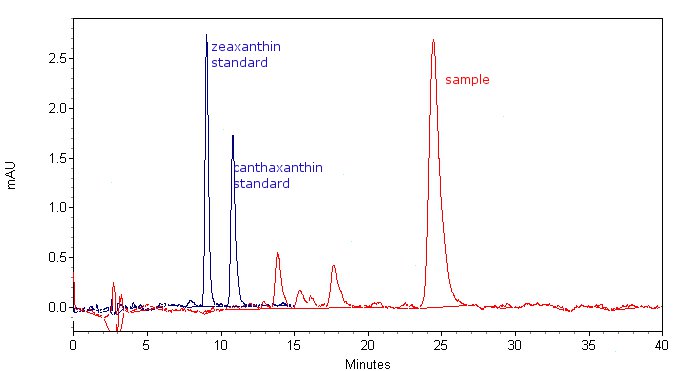
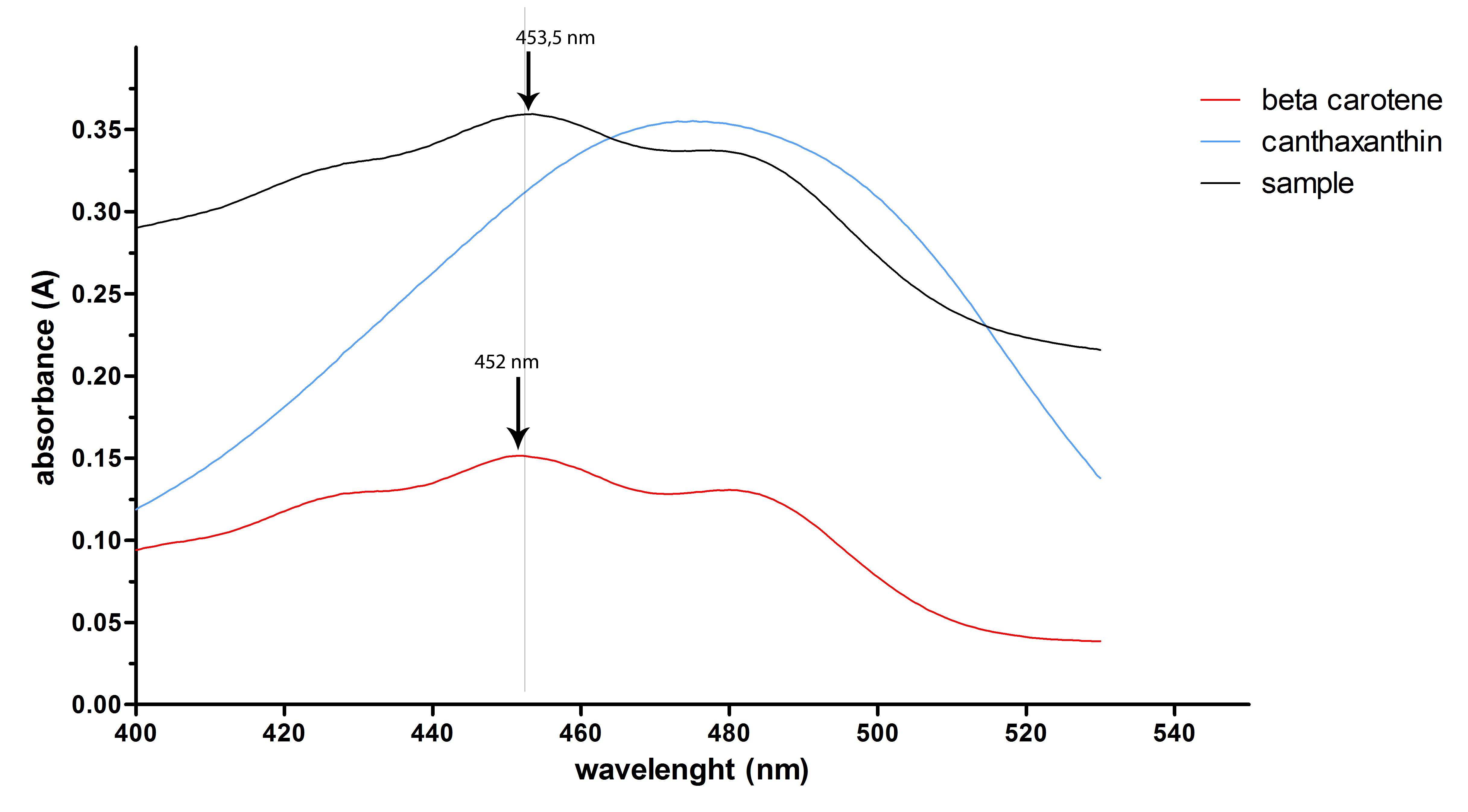
| - | + | We added a synthetic gene for the carotenoid biosynthetic enzyme (crtO) to the Registry. The enzyme crtO can introduce keto group to beta-carotene ring and could lead to formation of echinenone (one keto group) or canthaxanthine (two keto groups). We were not able to obtain standard for echinenone, but we conclude that it was probably present in the cell extract, as there was neither canthaxanthin or beta carotene in the sample (the latter is not present on the HPLC chromatogram, as it does not elute with the method used). There was, however, a coloured substance in the sample, with the absorbance spectrum characteristic for carotenoids, but with a slightly different absorbance maximum than beta carotene. | |
| - | + | ||
| - | + | [[Image:SLO_canthaxanthin_HPLC.jpg|thumb|center|700px|'''Figure 6:''' HPLC analysis of sample, extracted from cells containing R0011 crtEBIYO construct. Analysis shows the presence of a compound, which is less polar than zeaxanthin or canthaxanthin.]] | |
| - | + | ||
| - | + | [[Image:SLO_echinenon.jpg|thumb|center|700px|'''Figure 7:''' Absorbance spectra of the sample, extracted from cells containing R0011 crtEBIYO construct. The absorbance peak of the sample is slightly shifted to longer wavelenght in comparison to beta-carotene standard.]] | |
| - | + | ||
| - | + | [[Image:SLO_canthaxanthin_pathway.png|thumb|center|700px|'''Figure 8''' Biosynthesis of canthaxanthin from beta carotene. This is a two step reaction, the intermediate is an assymetric carotenoid called echinenone.]] | |
| - | + | ||
| - | + | <h2>DNA-guided assembly of carotenoid biosynthetic pathway</h2> | |
| - | + | ||
| - | < | + | We constructed 6 genes coding for zinc finger - carotenoid biosynthesis enzyme fusion proteins (Jazz::crtE, Blues::crtB, Zif268::crtI, PBSII::crtY, ZnF_HivC::crtO and Gli1::crtZ). Bacterial cells were transformed with three plasmids (two with each containing three enzymes and the third plasmid with DNA program). Therefore bacteria required three selection markers, which almost stopped their growth. Decreasing the amount of antibiotics allowed bacteria to grow faster but we could only demonstrate the production of lycopene, which can be produced by the first three enzymes. The reason is that probably bacteria lost a plasmid for the rest of the biosynthetic pathway. However production of lycopene shows that zinc fingers do not interfere with membrane enzyme activity. |
| - | + | ||
| - | + | [[Image:SLO_pelet_likopen2.JPG|thumb|center|350px|'''Figure 9:''' Cell pellets a culture with operon for lycopene biosynthesis with enzymes fused to zinc finger domains (pBAD Jazz::crtE Blues::crtB Zif268::crtI in pSB4K5)]] | |
| - | + | ||
| - | + | [[Image:SLO_HPLC_lycopene.jpg|thumb|center|700px|'''Figure 10:''' HPLC analysis of extract from cell pellet of a culture with operon for lycopene biosynthesis with enzymes fused to zinc finger domains (pBAD Jazz::crtE Blues::crtB Zif268::crtI in pSB4K5).]] | |
| - | + | ||
| - | + | <h2>Future perspectives</h2> | |
| - | + | ||
| - | + | We managed to prepare the constructs according to our cloning strategy. The whole system was on three different plasmids, which caused delayed growth of bacteria in the presence of three antibiotics. In order to obtain the expected results we will have to introduce the complete system for DNA-guided carotenoid biosystem into bacteria either in a single plasmid or vie chromosome integration. Due to the lack of time we did not manage to complete all of our goals, that is to extend the demonstration of the application of DNA-guided assembly of biosynthetic pathway from violacein also to carotenoids. Therefore the improvement of yield and specificity of carotenoid biosynthesis has yet to be determined in the next experiments. | |
| - | + | ||
| - | <h2 | + | <hr> |
| - | + | ||
| - | + | http://www.bccresearch.com/report/FOD025C.html, 26.10.2010<br><br> | |
| - | + | Scaife, M.a., Burja, A.M. & Wright, P.C., 2009. Characterization of cyanobacterial beta-carotene ketolase and hydroxylase genes in Escherichia coli, and their application for astaxanthin biosynthesis. <em>Biotechnology and bioengineering</em>, 103(5), 944-55. | |
| - | <hr | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | < | + | |
<!--STOP BESEDILO--> | <!--STOP BESEDILO--> | ||
Latest revision as of 01:13, 28 October 2010
Contents |
State of the art
- Carotenoid biosynthetic pathway available in the Registry demonstrated production of carotenoids up to beta-carotene
Aims
- Add additional genes enzymes and achieve biosynthesis of carotenoids zeaxanthin and canthaxanthin
- Prepare DNA-guided biosynthetic pathways for production of zeaxanthin and canthaxanthin in E.coli
Achievements
- We demonstrated production of zeaxanthin in E.coli using parts from the Registry
- We added an additional carotenoid biosynthetic pathway enzyme crtO and demonstrated formation of a carotenoid that was probably echinenone
- We constructed DNA-guided biosynthetic pathway constructs but did not have time to test it experimentally as the cells with three plasmids grow extremely slow
Background
Carotenoids form a large family of colored compounds, naturally occuring in plants and bacteria. Besides being vitamin A precursors, many carotenoids have a potent antioxidant activity and are therefore widely used as food supplements, food colorants in cosmetics and as animal feed. According to BCC Research total global market for carotenoids was worth $766 million in 2007 and is expected to increase to $909 million till 2015. The largest share goes to beta carotene, with asthaxanthin closely following and canthaxanthin in the third place.
Due to difficulties in obtaining some carotenoids from natural sources on a large scale, they are produced by chemical synthesis. But especially in food industry they are generally preferred from natural sources over synthetic compounds. Also due to the cost and sustainability, there is a large interest to use microbial hosts for carotenoid biosynthesis.
Biosynthetic pathway
Many carotenoids have been produced in E.coli using reconstitution of a plant biosynthetic pathway in bacteria. The precursor for carotenoids is farnesyl pyrophoshpate. The first three enzymes in the biosynthetic pathway produce lycopene, a red pigment found in tomatoes. The next enzyme in the pathway, lycopene beta-cyclase (crtY), can transform it to beta carotene, which is a vitamin A precursor. This segment of the carotenoid biosynthetic pathway was already available in the Registry and iGEM teams from previous years (Edinburgh 07, Edinburgh 08, Guelph 08, Cambridge09) demonstrated production of beta-carotene and lycopene.
Beta carotene can be further oxidised by the enzyme beta hydroxylase (crtZ), which results in zeaxanthin, a yellow pigment found in bell peppers or corn and occuring in retina of the eye. This is the reason that zeaxanthin intake is essential to prevent eye disorders, such as age-related mascular degeneration as the major cause of visual impairment in older adults, glaucoma or cataract. The gene coding crtZ was also avaliable in the Registry but its funcion has not yet been demonstrated.
Another biosynthetic pathway of beta carotene modification is oxidation by the enzyme beta ketolase (crtO), which results in another industrially interesting compound - canthaxanthin. This enzyme, however, preferrably oxidizes only one ring of the beta carotene, which leads to formation of carotenoid called echinenone, but when the gene is overexpressed, canthaxanthin can be produced as well. Canthaxanthin can also be further modified by beta hydroxylase, which gives red pigment asthaxanthin as a product.
Aims
We wanted to improve the velocity, yield and specificity of carotenoid biosynthesis in E. coli using the DNA-guided biosynthetic enzyme ordered assembly. We constructed genes coding for chimeric enzyme - zinc finger fusion proteins (see scheme below) and designed two DNA programs, which specifically assemble carotenoid biosynthetic enzymes of each pathway. A sequence of crtE, crtB, crtI, crtY, crtO and crtZ enzymes should produce asthaxanthin, while crtE, crtB, crtI, crtY and crtZ should result in the production of zeaxanthin. (see scheme below).
One of the reasons why carotenoid biosynthesis pathway represented a challenge for us was putative membrane localisation of at least a few enzymes of the pathway. The literature does not provide the answer to this question. Membrane localization of some enzymes would mean that some enzymes are in a way already compartmentalized in the cell, but we nevertheless expected some improvement using DNA guided assembly. Another question regarding the membrane localization of the enzymes was whether they will retain their activity if fused to zinc finger domain, as additional domain at the N-terminus could disturb their membrane association.
Cloning strategy
As the sum of lengths of genes coding for chimeric enzymes for carotenoid biosynthesis is almost 9 kbp, we decided to clone them into two separate plasmids with different antibiotic resistance. We cloned the first three genes (Jazz::crtE, Blues::crtB, Zif268::crtI) into a vector with kanamycin resistance (pSB4K5) and the last three (PSBII::crtY, ZnfHivC::crtO and crtZ::Gli1) into a vector with chloramphenicol resistance (pSB4C5).
To faciliate the cloning process we designed our chimeric enzymes with zinc finger domain N-terminal to the enzymes. The genes for enzymes from The Registry include both start and stop codon, which meant we could directly fuse them to C-terminus of our synthetic zinc finger domains which do not have stop codon.
We chose a linker olygopeptide of 10 amino acids, consisting of Gly and Ser residues to connect zinc fingers and enzymes, because it enables the flexibility and has also been used for similar purposes before. DNA program encoding the sequence of chimeric enzymes was introduced into bacteria on the third plasmid.
Results
Biosynthesis of zeaxanthin
We demonstrated that enzyme crtZ can produce zeaxanthin from beta carotene by inserting the gene, coding for this enzyme, behind the beta carotene-operon, submitted to Registry of parts by teams from previous years. This extends the verified carotenoid biosynthetic pathway in E.coli available in the Registry for an additional step.
Biosynthesis of canthaxanthin
We added a synthetic gene for the carotenoid biosynthetic enzyme (crtO) to the Registry. The enzyme crtO can introduce keto group to beta-carotene ring and could lead to formation of echinenone (one keto group) or canthaxanthine (two keto groups). We were not able to obtain standard for echinenone, but we conclude that it was probably present in the cell extract, as there was neither canthaxanthin or beta carotene in the sample (the latter is not present on the HPLC chromatogram, as it does not elute with the method used). There was, however, a coloured substance in the sample, with the absorbance spectrum characteristic for carotenoids, but with a slightly different absorbance maximum than beta carotene.
DNA-guided assembly of carotenoid biosynthetic pathway
We constructed 6 genes coding for zinc finger - carotenoid biosynthesis enzyme fusion proteins (Jazz::crtE, Blues::crtB, Zif268::crtI, PBSII::crtY, ZnF_HivC::crtO and Gli1::crtZ). Bacterial cells were transformed with three plasmids (two with each containing three enzymes and the third plasmid with DNA program). Therefore bacteria required three selection markers, which almost stopped their growth. Decreasing the amount of antibiotics allowed bacteria to grow faster but we could only demonstrate the production of lycopene, which can be produced by the first three enzymes. The reason is that probably bacteria lost a plasmid for the rest of the biosynthetic pathway. However production of lycopene shows that zinc fingers do not interfere with membrane enzyme activity.
Future perspectives
We managed to prepare the constructs according to our cloning strategy. The whole system was on three different plasmids, which caused delayed growth of bacteria in the presence of three antibiotics. In order to obtain the expected results we will have to introduce the complete system for DNA-guided carotenoid biosystem into bacteria either in a single plasmid or vie chromosome integration. Due to the lack of time we did not manage to complete all of our goals, that is to extend the demonstration of the application of DNA-guided assembly of biosynthetic pathway from violacein also to carotenoids. Therefore the improvement of yield and specificity of carotenoid biosynthesis has yet to be determined in the next experiments.
http://www.bccresearch.com/report/FOD025C.html, 26.10.2010
Scaife, M.a., Burja, A.M. & Wright, P.C., 2009. Characterization of cyanobacterial beta-carotene ketolase and hydroxylase genes in Escherichia coli, and their application for astaxanthin biosynthesis. Biotechnology and bioengineering, 103(5), 944-55.
 "
"