Results
Photosensor
Biobrick assembly and sequence
We succesfully assembled the biobrick K343007, which consists of the TetR repressible promoter R0040 followed by the ribosome binding site B0034, the coding sequence K343003 and the terminators B0010 and B0012.
This was achieved through "standard" silver assembly and confirmed via sequencing with biobrick verification forward 2 (VF2) and verification reverse (VR) primers.
Sequencing results (as .ab1 files):
Sequencing results K343007
Effect
Stability assay
| Stability assay. |
|
|
|
|
|
|
|
|
|
| 0
generations |
Dilution number |
Colonies counted on
plate |
Cfu |
PS-chlor/PS+chlor |
| PS1 +
chlor |
6 |
168 |
8.4E+09 |
100 |
| PS1 -
chlor |
6 |
157 |
7.85E+09 |
|
| PS2 +
chlor |
6 |
192 |
9.6E+09 |
100 |
| PS2 -
chlor |
6 |
216 |
1.08E+10 |
|
| 20
generations |
|
|
|
|
| PS1 +
chlor |
6 |
157 |
7.85E+09 |
59.24528302 |
| PS1 -
chlor |
6 |
265 |
1.325E+10 |
|
| PS2 +
chlor |
6 |
130 |
6.5E+09 |
63.41463415 |
| PS2 -
chlor |
6 |
205 |
1.025E+10 |
|
| 40
generations |
|
|
|
|
| PS1 +
chlor |
6 |
143 |
7.15E+09 |
95.97315436 |
| PS1 -
chlor |
6 |
149 |
7.45E+09 |
|
| PS2 +
chlor |
6 |
168 |
8.4E+09 |
92.81767956 |
| PS2 -
chlor |
6 |
181 |
9.05E+09 |
|
| 60
generations |
|
|
|
|
| PS1 +
chlor |
6 |
147 |
7.35E+09 |
89.63414634 |
| PS1 -
chlor |
6 |
164 |
8.2E+09 |
|
| PS2 +
chlor |
6 |
134 |
6.7E+09 |
104.6875 |
| PS2 -
chlor |
6 |
128 |
6.4E+09 |
|
| 80
generations |
|
|
|
|
| PS1 +
chlor |
6 |
127 |
6.35E+09 |
136.5591398 |
| PS1 -
chlor |
6 |
93 |
4.65E+09 |
|
| PS2 +
chlor |
6 |
99 |
4.95E+09 |
104.2105263 |
| PS2 -
chlor |
6 |
95 |
4.75E+09 |
|
| 100
generations |
|
|
|
| PS1 +
chlor |
5 |
130 |
650000000 |
125 |
| PS1 -
chlor |
5 |
104 |
520000000 |
|
| PS2 +
chlor |
5 |
124 |
620000000 |
118.0952381 |
| PS2 -
chlor |
5 |
105 |
525000000 |
|
| Number of generations |
PS1 |
PS2 |
PS
(average) |
Standard
deviations |
| 0 |
100 |
100 |
100 |
0 |
| 20 |
59.24528 |
63.41463 |
61.32995858 |
2.948176455 |
| 40 |
95.97315 |
92.81768 |
94.39541696 |
2.231257632 |
| 60 |
89.63415 |
104.6875 |
97.16082317 |
10.64432845 |
| 80 |
136.5591 |
104.2105 |
120.3848331 |
22.87392395 |
| 100 |
125 |
118.0952 |
121.547619 |
4.882403965 |
Flagella
Motility assay
Exp. 1
The purpose of this experiment is to see if the cells containing pSB1C3-K343004 move farther than the wild type (MG1655) and the negative control (DH5alpha). This would be an indication of hyperflagellation. We also want to test if it makes a difference in the motility wether the bacteria contain low-medium- or high-copy plasmids.
For the motility experiments we added 5ul of an ON culture to petridishes containing motility agar (LB media with 0.3% agar) instead of regular LA (Luria agar). This semi-solid media lets the bacteria swim more easily.
The plates were incubated at 37 degrees celcius for 24 hours.
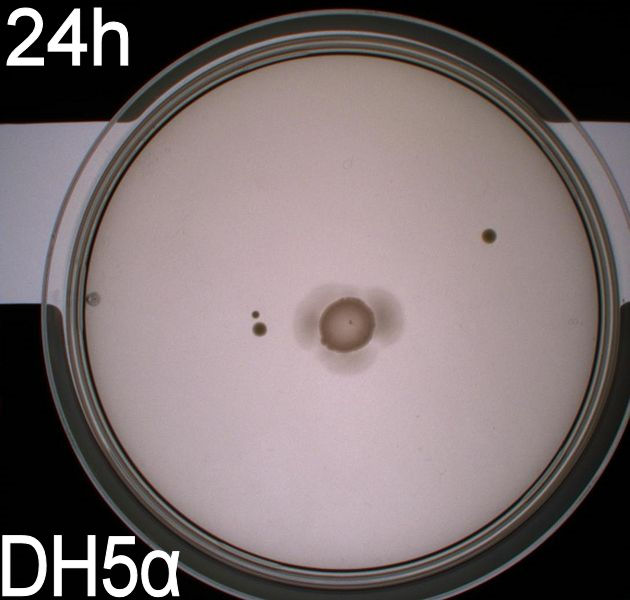



The upper two plates did not contain antibiotics, and therefore contamination colonies are seen.
The upper left picture is of E. coli strain DH5alpha that does not express flagella and therefor movement in the media should, as seen on the picture, be minimal. The upper right picture is of the wild type E. coli strain MG1655, this strain has about 8-10 flagella per cell. These cells are, as seen, expected to move farther than the DH5alpha but not as far as the transformed cells. The lower left picture is of E. Coli strain MG1655 with pSB1C3-K343004 this shows that these bacteria move farther than the wild type and the negative control. The lover right picture is of E. coli strain MG1655 with pSB3K3-K343004 these bacteria move farther than the wild type, the negative control and MG1655 with pSB3K3-K343004.
From the pictures above we can definately se that the bacteria containing our part is much more motile than the wild type. We assume this is caused by overexpression of the FlhDC master flagella operon which leads to hyperflagellation of the cells.
The two buttom pictures show that bacteria with pSB1C3-K343004 have not moved as far as the bacteria containing pSB3K3-K343004. pSB1C3 is a high copy plasmid while pSB3K3 is a low-medium copy plasmid. The promoters in K343004 is a constitutive promoter (tetR repressable promoter). Bacteria containing a high copy plasmid with a constitutive promoter are more metabolically challanged than bacteria containing a low- or medium-copy plasmid with a constitutive promoter because of the higher number of plasmids per the cell. Therefore the high copy plasmid bacteria are less motile than low- or medium-copy plasmid bacteria.
Retinal
UV-vis determination of beta-carotene production
In this experiment cells were prepared and harvested according to protocol [1]. This experiment was performed with four different strains of E. coli:
Wildtype Top10
Wildtype MG1655
Top10 containing PSB1A2 with constitutively active K274210
MG1655 containing PSB1A2 with constitutively active K274210
The measurements were performed on cells both in the exponential and stationary phase. The resulting graphs are presented below:
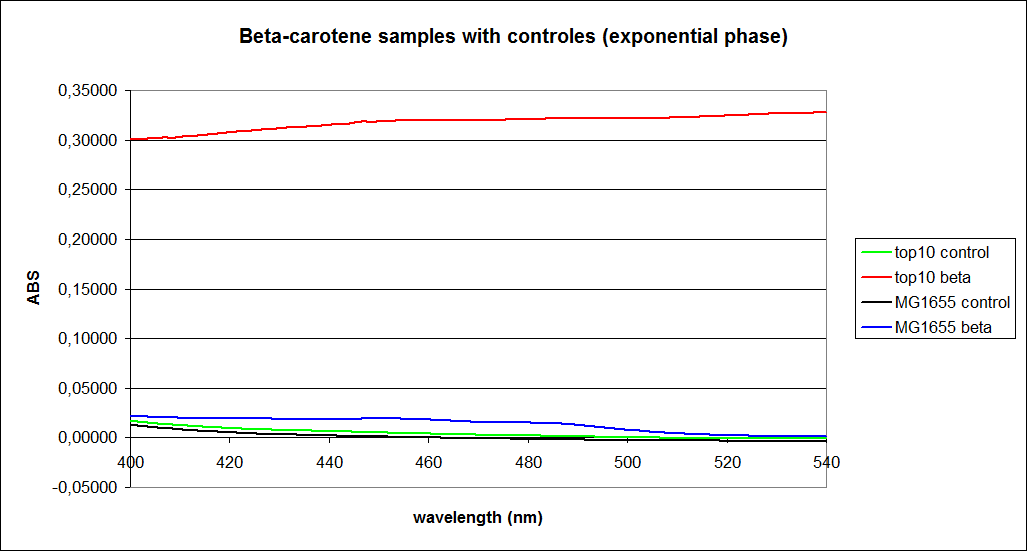
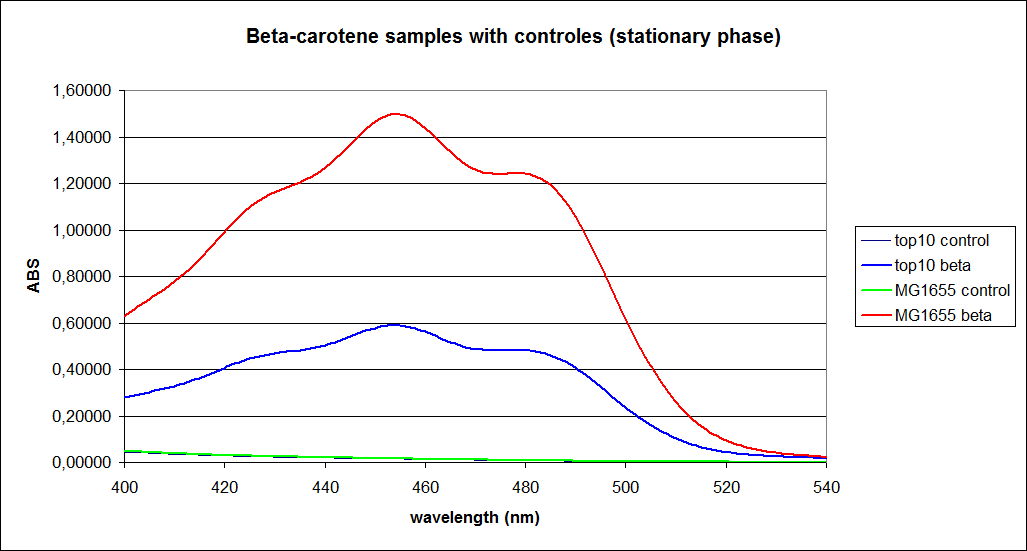
In order to assess the results, a series of standard dilutions of beta-carotene were made. The concentrations were 1mM, 100 µM, 50 µM, 25 µM, 10 µM, 5 µM, 1 µM, 100 nM and 10 nM. The standard dilutions were also measured on the spectrophotometer. The resulting graphs are presented low:
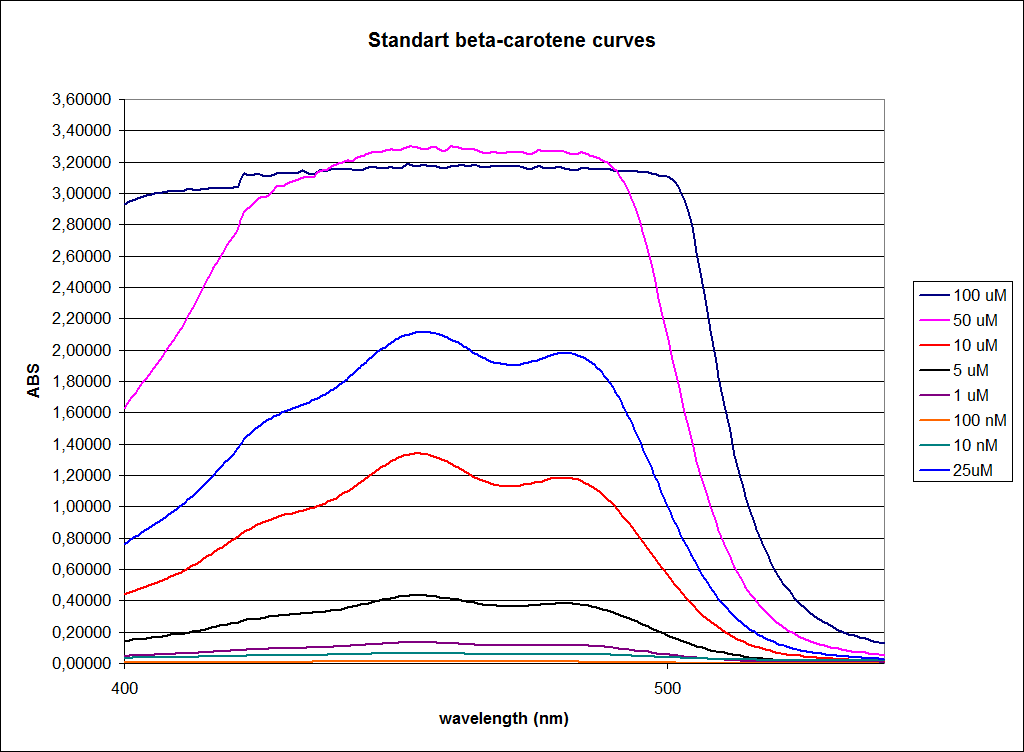
When combining the graphs of the samples and the standard dilutions, it is clear that the spectres are uniform. This similarity of the spectres indicates that the cells actually do produce beta-carotene. The graphs also give a qualitative indication of the amount of beta-carotene produced by the MG1655 and the Top10 E. coli cells. The resulting graphs are presented below:
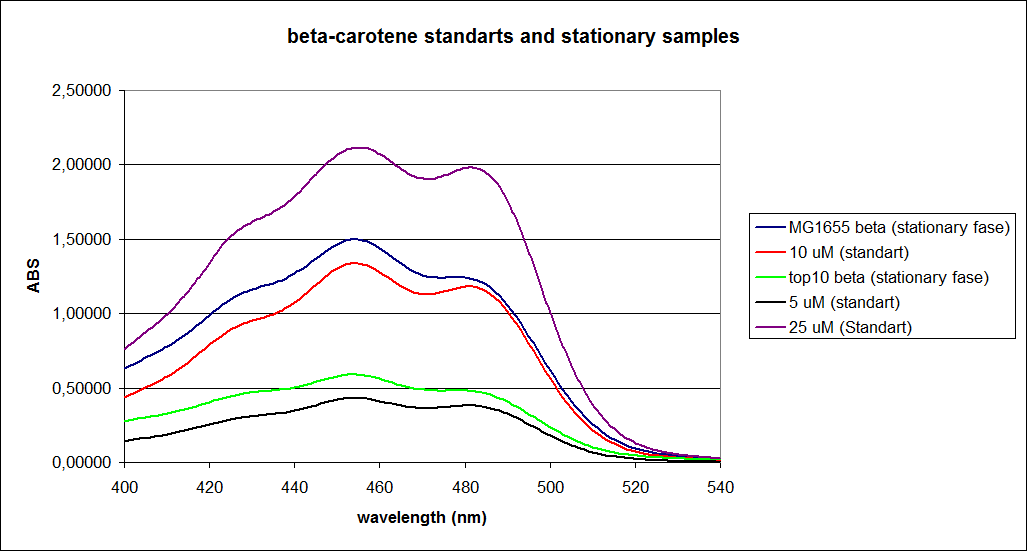
UV-Vis determination of beta-carotene and retinal production
In this experiment cells was prepare and harvested according to protocol [2]. This experiment was preformed with six different strain of E. Coli:
wildtype Top10
wildtype MG1655
Top10 containing PSB1A2 with K274210 constitutively active
MG1655 containing PSB1A2 with K274210 constitutively active
TOP10 containing PSB1A2 with K274210 plus PSB1C3 with K343005 both under a constitutively active promotor
MG1655 containing PSB1A2 with K274210 plus PSB1C3 with K343005 both under a constitutively active promotor
The measurements were preformed on cells after 20 hours of growth, because the first experiment showed none or little product produced in the exponential phase. The resulting graphs is presented beneath the text.
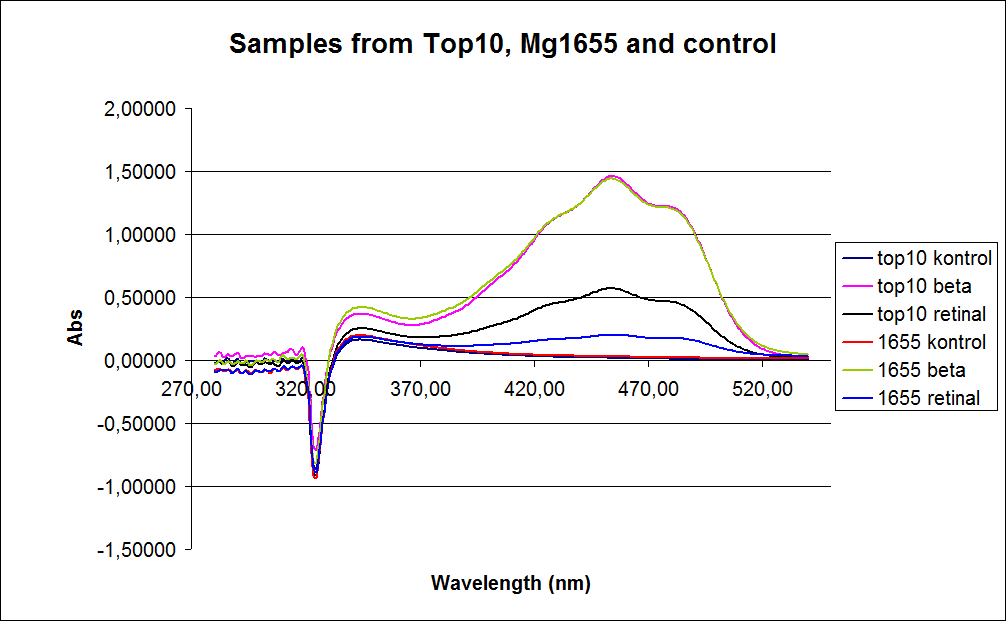
In the spectre a sudden drop at 320 nm occurs, this cannot be explained .
The graph also shows that some cell material interferes with the UV-Vis measurement at the retinal has the strongest absorption, because of this some organic separation might be needed before the measurement is preformed.
In order assess the results a standard dilution of beta-carotene and retinal was made and measured: 1mM, 100 µM, 50 µM, 25 µM, 10 µM, 5 µM, 1 µM, 100 nM and 10 nM. The standart dilutions was also measured on the spectrophotometer. The resulting graphs is presented beneath the text
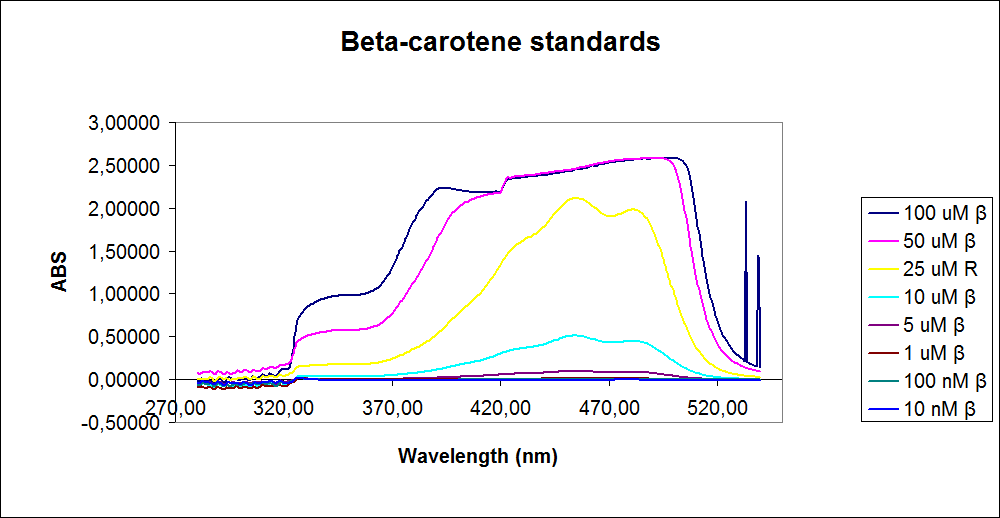
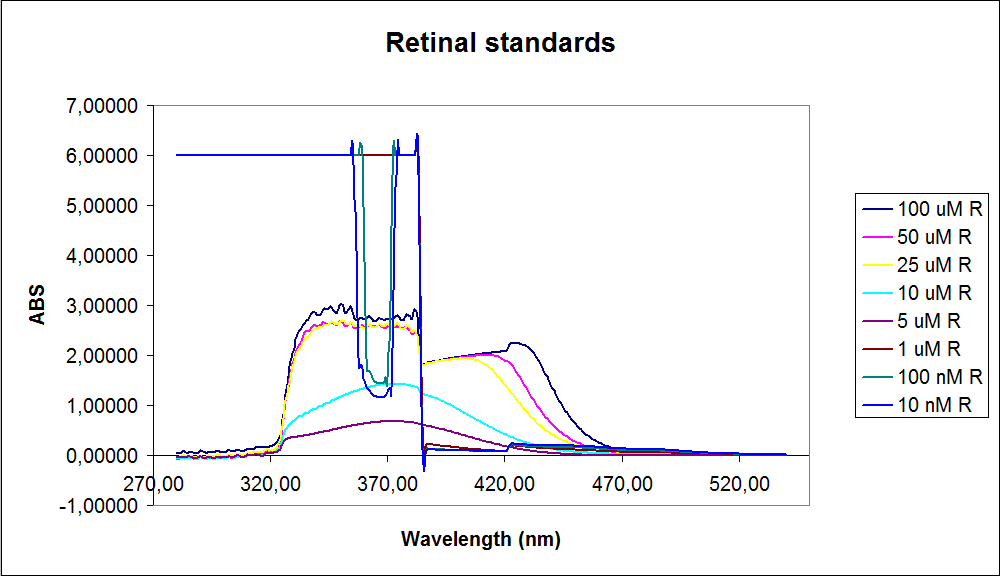
The spectrum for the Beta-carotene looks normal and reliable, the standards for retinal however doesn’t look reliable. The spectrum for the three lowest concentrations appears to be give a wrong picture from the measurement.
The measurement jumps rapidly from nearly nothing to the maximum absorbance, this might be because the concentration of retinal is to low to be measured as a spectrum, but the concentration of the individual derivates of retinal might be high enough to be measured.
It might also just be that the concentrations are to low and something ells are influencing the measurements.
Because of the error in the UV-vis spectre concerning retinal detection in both the standards and samples the next characterization experiments is preformed on a HPLC.
OD measurment of MG1655
|
Mg1655 |
|
PS in pSB3T5 |
PS in pSB1C3 |
flhDC in pSB3K3 |
|
flhDC
in pSB1C3 |
|
NinaB in pSB1C3 |
| Hours |
1 |
2 |
1 |
2 |
1 |
2 |
1 |
2 |
1 |
2 |
1 |
2 |
| 0 |
0.024 |
0.026 |
0.028 |
0.03 |
0.024 |
0.024 |
0.028 |
0.027 |
0.028 |
0.027 |
0.027 |
0.027 |
| 1 |
0.082 |
0.03 |
0.058 |
0.06 |
0.059 |
0.063 |
0.069 |
0.077 |
0.052 |
0.051 |
0.084 |
0.072 |
| 2.5 |
0.58 |
0.65 |
0.42 |
0.45 |
0.48 |
0.44 |
0.52 |
0.6 |
0.38 |
0.34 |
0.31 |
0.54 |
| 3 |
1.14 |
1.16 |
0.99 |
0.91 |
0.94 |
0.94 |
1.06 |
1.16 |
0.84 |
0.78 |
1.04 |
1.03 |
| 3.83 |
1.54 |
1.66 |
1.52 |
1.31 |
1.38 |
1.37 |
1.54 |
1.72 |
1.33 |
1.25 |
1.91 |
1.45 |
| 5 |
4.4 |
4.3 |
3.2 |
3.4 |
4.2 |
3.3 |
4.2 |
2.2 |
3.9 |
4 |
3.9 |
3.9 |
| 6 |
4.7 |
4.7 |
2.8 |
4.3 |
4.2 |
3.1 |
4.7 |
4.4 |
3.8 |
5 |
3.4 |
4.7 |
| 7 |
4 |
3.8 |
3.3 |
3.8 |
4.2 |
3.2 |
4.3 |
4.7 |
4.9 |
4.5 |
3.7 |
3.9 |
| 8 |
5.5 |
6.1 |
3.9 |
4 |
3.5 |
6.8 |
4.2 |
4.8 |
5.4 |
5.3 |
4.6 |
4.6 |
| 9 |
4.1 |
4.1 |
3.4 |
2.6 |
3.6 |
4.7 |
2.6 |
3.1 |
3.7 |
3.1 |
2.6 |
3.6 |
| 10 |
4.6 |
6.1 |
5 |
4.8 |
7.8 |
4.5 |
5 |
6 |
5.3 |
5.3 |
4.4 |
4.7 |
| 11 |
5.1 |
6.2 |
4.7 |
4 |
6.1 |
7.1 |
6.6 |
8.3 |
6.6 |
6.4 |
6.2 |
4.1 |
| 12 |
5.1 |
5 |
3 |
4.4 |
4.4 |
3.9 |
4 |
4.8 |
5 |
5.3 |
4 |
4.7 |
| 24 |
5.2 |
5.6 |
3.3 |
5.4 |
6.4 |
5.8 |
4.6 |
6.4 |
7.1 |
7.4 |
5.6 |
5.2 |
 "
"










