Team:SDU-Denmark/project-r
From 2010.igem.org
(Difference between revisions)
m (→Video microscopy results) |
m (→Results) |
||
| Line 9: | Line 9: | ||
== Photosensor == | == Photosensor == | ||
=== Motility assay === | === Motility assay === | ||
| + | <p style="text-align: justify;"> | ||
<br> | <br> | ||
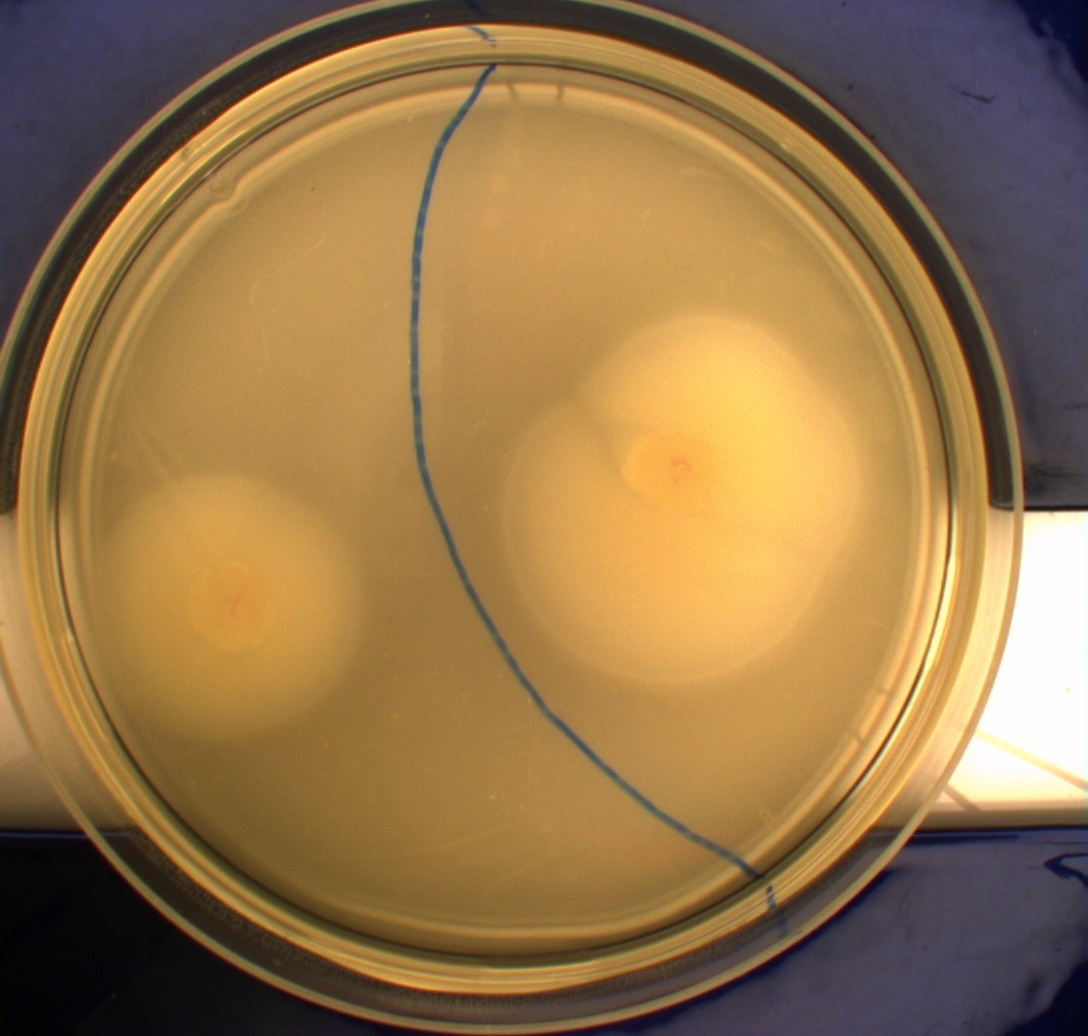
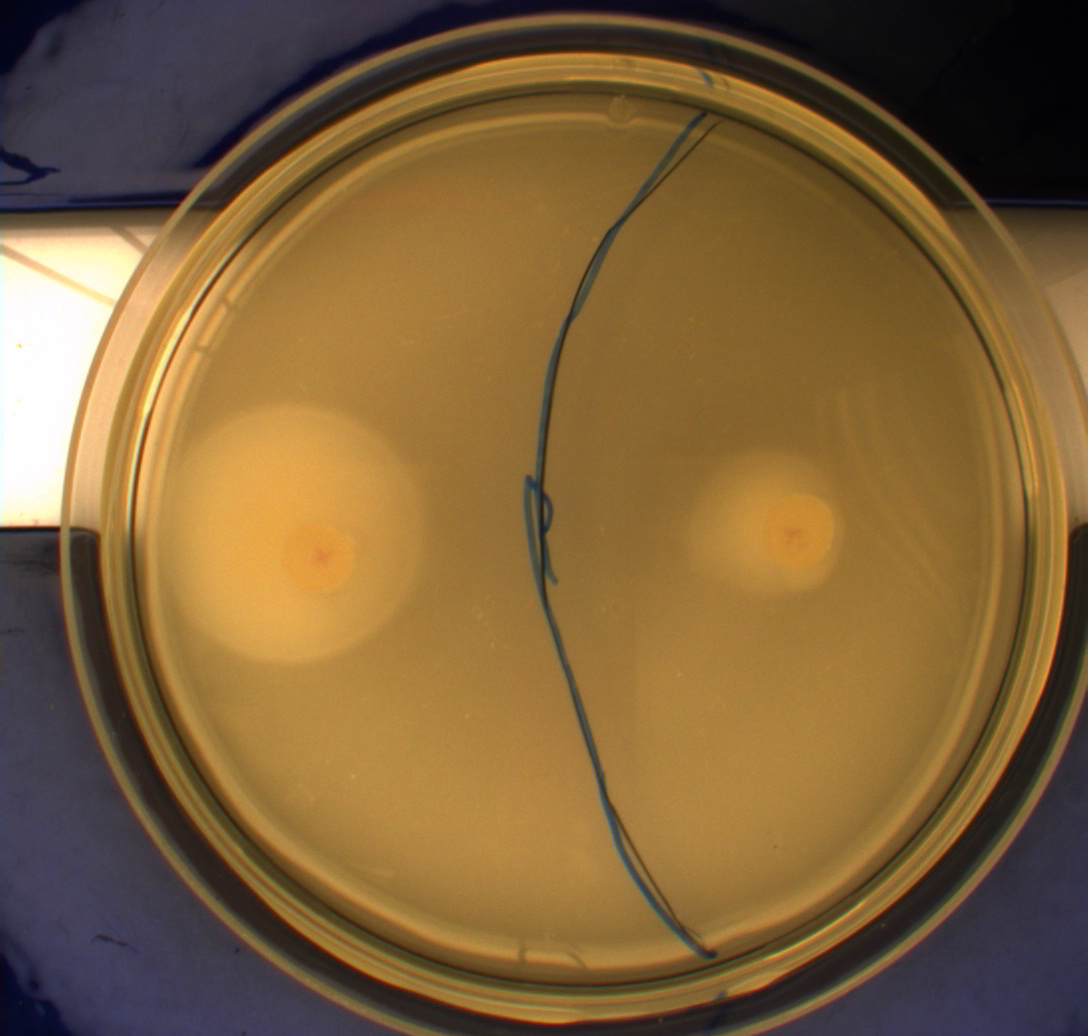
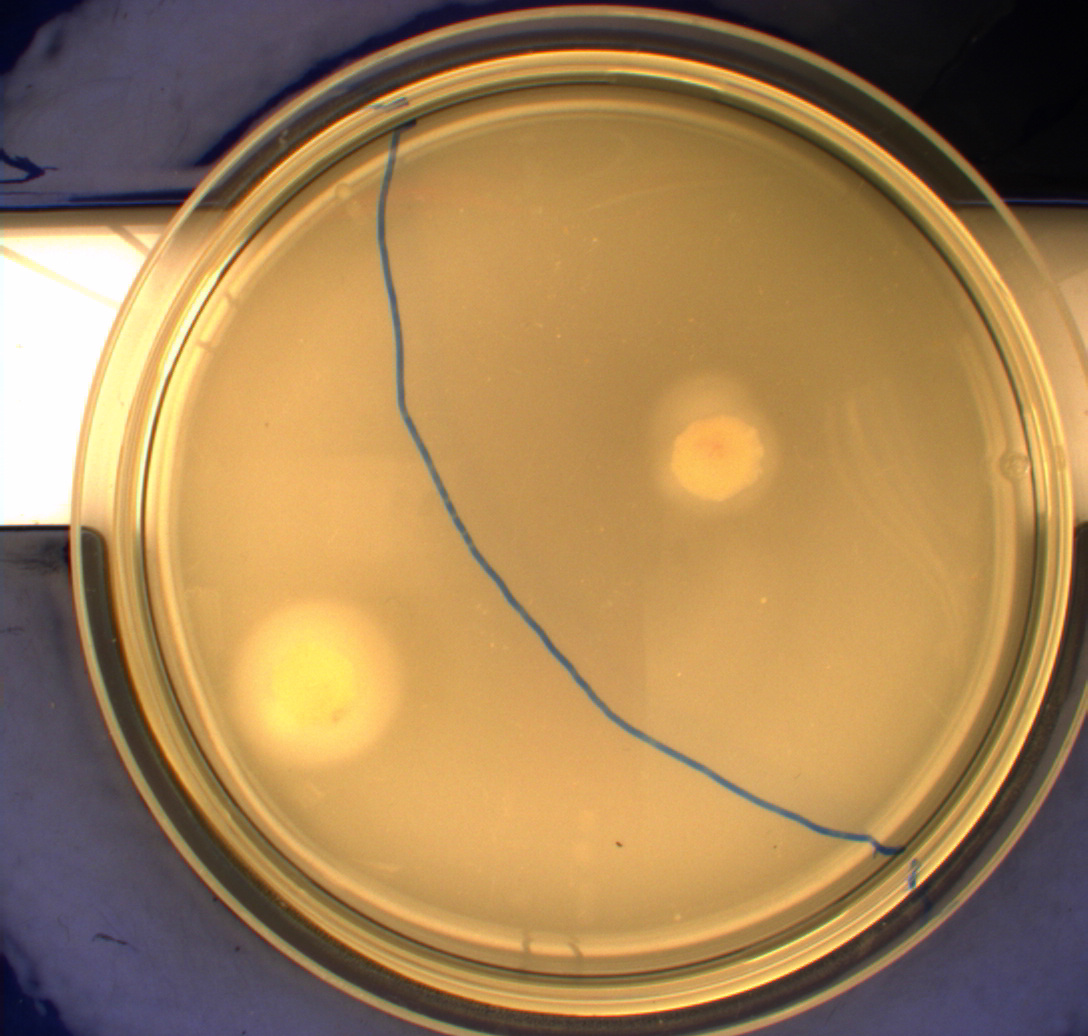
In this experiment, we shone blue light on one half of the plate while the other half was in the dark. We then placed one colony in each half and observed their motility pattern after 24 hours. We did this for three different strains of ''E. coli'': DH5alpha, Wildtype Mg1655 and Mg1655 containing a plasmid with our photosensor constitutively on. See the results for yourself: Light shone on the right half of the plates, the left half was in the dark.<br> | In this experiment, we shone blue light on one half of the plate while the other half was in the dark. We then placed one colony in each half and observed their motility pattern after 24 hours. We did this for three different strains of ''E. coli'': DH5alpha, Wildtype Mg1655 and Mg1655 containing a plasmid with our photosensor constitutively on. See the results for yourself: Light shone on the right half of the plates, the left half was in the dark.<br> | ||
| Line 21: | Line 22: | ||
This means that if the photosensor leads to a reduced tumbling rate when exposed to blue light, it should look a lot like the colony of DH5alpha which was exposed to blue light. This is exactly the case which proves that the photosensor has an effect on the tumbling rate of the bacteria. The photosensor colony in the dark behaved like the Mg1655 colony in the dark and spread out. This is normal as cells that do not tend to either tumble or run a lot, will spread the most. <br><br> | This means that if the photosensor leads to a reduced tumbling rate when exposed to blue light, it should look a lot like the colony of DH5alpha which was exposed to blue light. This is exactly the case which proves that the photosensor has an effect on the tumbling rate of the bacteria. The photosensor colony in the dark behaved like the Mg1655 colony in the dark and spread out. This is normal as cells that do not tend to either tumble or run a lot, will spread the most. <br><br> | ||
| - | + | </p> | |
=== Video microscopy results === | === Video microscopy results === | ||
| + | <p style="text-align: justify;"> | ||
<br> | <br> | ||
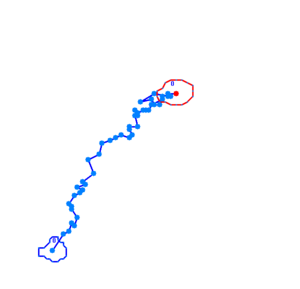
Bacteria containing the photosensor will exhibit a lowered tumbling rate when exposed to blue light (wavelengths around 350nm - 450nm). This was analysed with the help of video microscopy and the open source software [http://db.cse.ohio-state.edu/CellTrack/ "CellTrack"]. The individual cells' trajectory was tracked and their speed measured. The tracking results are as follows: | Bacteria containing the photosensor will exhibit a lowered tumbling rate when exposed to blue light (wavelengths around 350nm - 450nm). This was analysed with the help of video microscopy and the open source software [http://db.cse.ohio-state.edu/CellTrack/ "CellTrack"]. The individual cells' trajectory was tracked and their speed measured. The tracking results are as follows: | ||
| Line 30: | Line 32: | ||
[[Image:Team-SDU-Denmark-WTblue1.png |250px|Wildtype bacteria exposed to blue light]]<br> | [[Image:Team-SDU-Denmark-WTblue1.png |250px|Wildtype bacteria exposed to blue light]]<br> | ||
The phototaxic bacteria move more in a straight line when exposed to bluelight, as can be seen when comparing the trajectories of the three bacteria given earlier. These were taken from a batch of 10 cells tracked per sample. | The phototaxic bacteria move more in a straight line when exposed to bluelight, as can be seen when comparing the trajectories of the three bacteria given earlier. These were taken from a batch of 10 cells tracked per sample. | ||
| - | + | </p> | |
== Retinal == | == Retinal == | ||
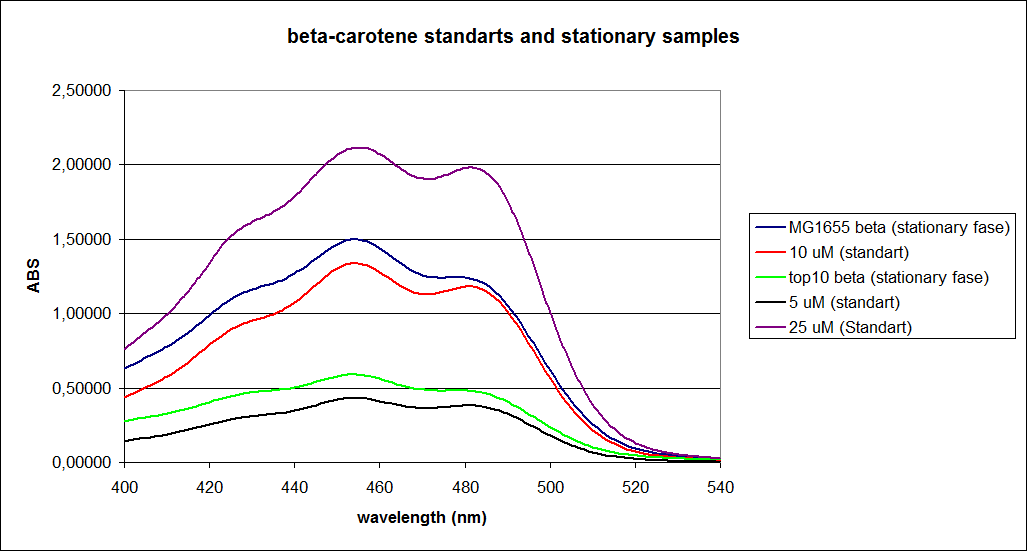
=== UV-vis spectrophotometer determination of beta-carotene production === | === UV-vis spectrophotometer determination of beta-carotene production === | ||
| + | <p style="text-align: justify;"> | ||
<br> | <br> | ||
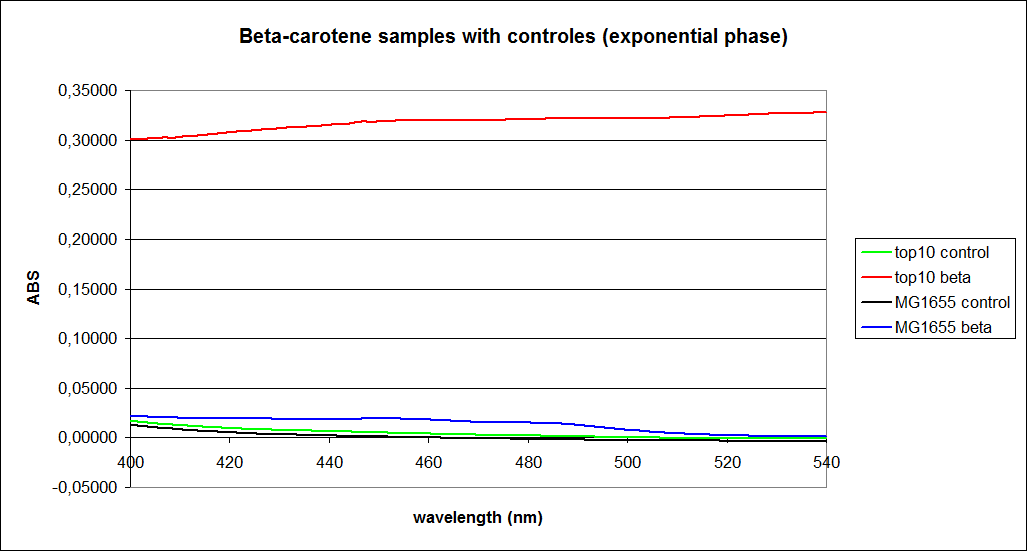
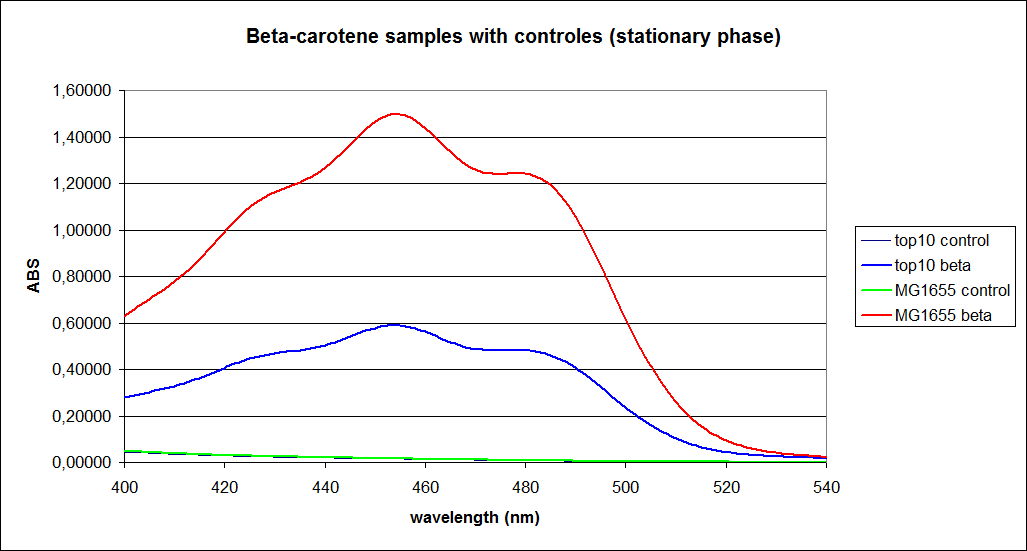
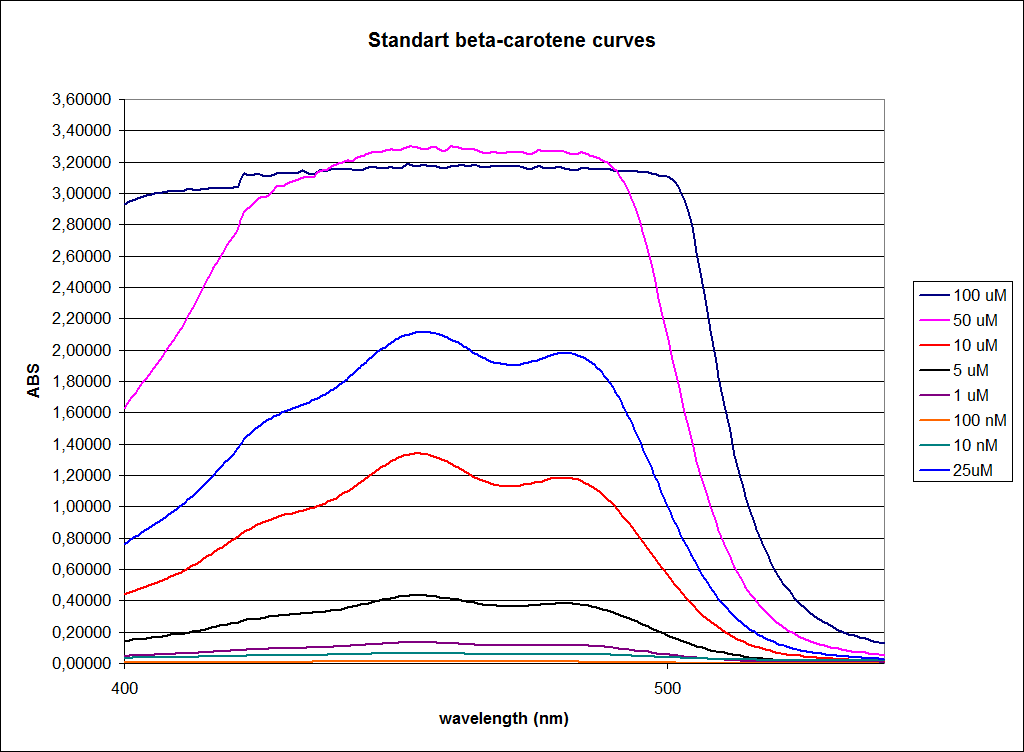
In this experiment cells were prepared and harvested according to protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#EX1.1]. This experiment was performed with four different strains of ''E. coli'': <br> | In this experiment cells were prepared and harvested according to protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#EX1.1]. This experiment was performed with four different strains of ''E. coli'': <br> | ||
| Line 54: | Line 57: | ||
[[Image:Image-Team-SDU-denmarkBetacarotene standarts and stationary samples.png |350px]] | [[Image:Image-Team-SDU-denmarkBetacarotene standarts and stationary samples.png |350px]] | ||
<br><br> | <br><br> | ||
| - | + | </p> | |
<br> | <br> | ||
http://igem.sdu.dk/wp-content/uploads/sponsor-sdu.png http://igem.sdu.dk/wp-content/uploads/sponsor-fermentas.png http://igem.sdu.dk/wp-content/uploads/sponsor-dnatech.png [[Image:Team-sdu-2010-ida-logo.png]]<br> | http://igem.sdu.dk/wp-content/uploads/sponsor-sdu.png http://igem.sdu.dk/wp-content/uploads/sponsor-fermentas.png http://igem.sdu.dk/wp-content/uploads/sponsor-dnatech.png [[Image:Team-sdu-2010-ida-logo.png]]<br> | ||
Revision as of 11:08, 21 October 2010
 "
"