Team:SDU-Denmark/project-r
From 2010.igem.org
(Difference between revisions)
m (→UV-Vis spectrophotometer determination of beta-carotene production) |
m (→Motility assay) |
||
| Line 10: | Line 10: | ||
=== Motility assay === | === Motility assay === | ||
<br> | <br> | ||
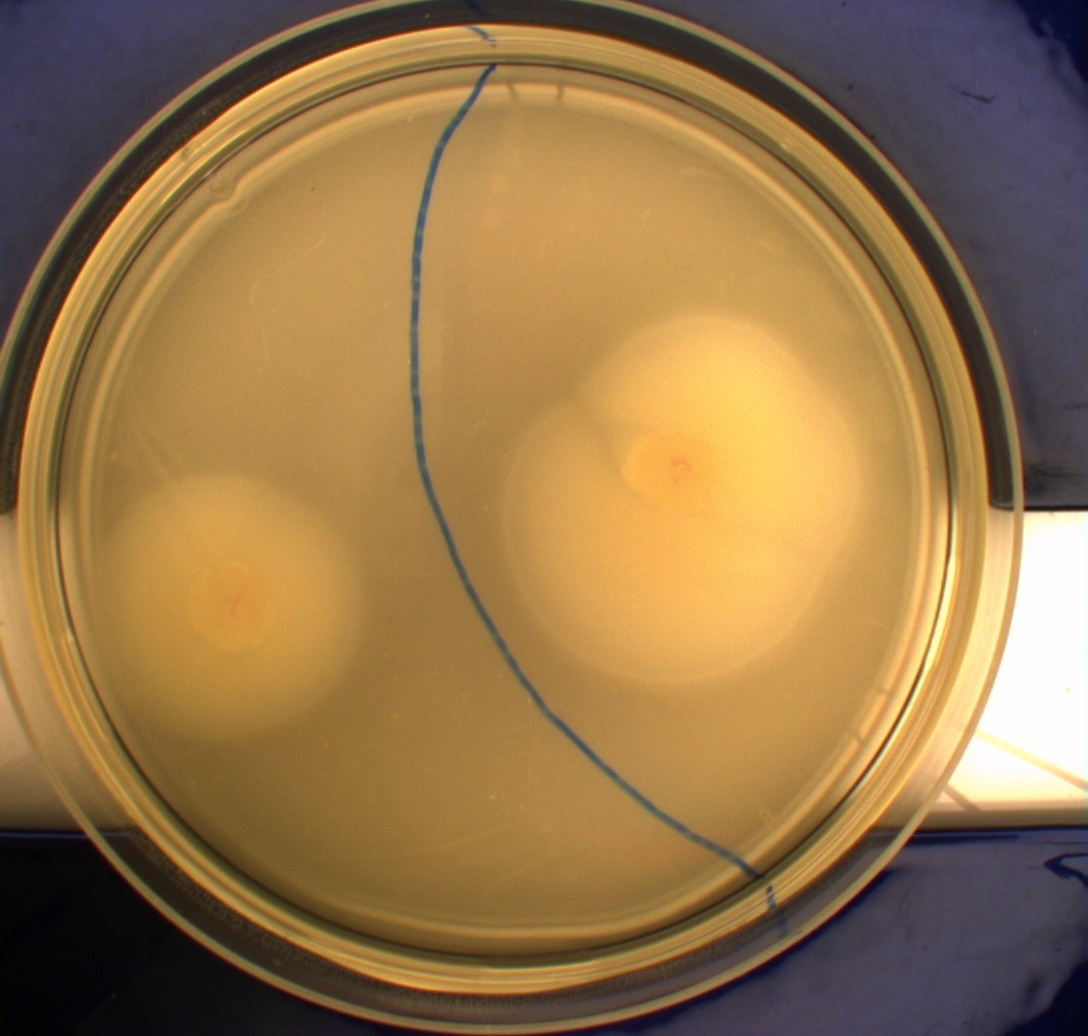
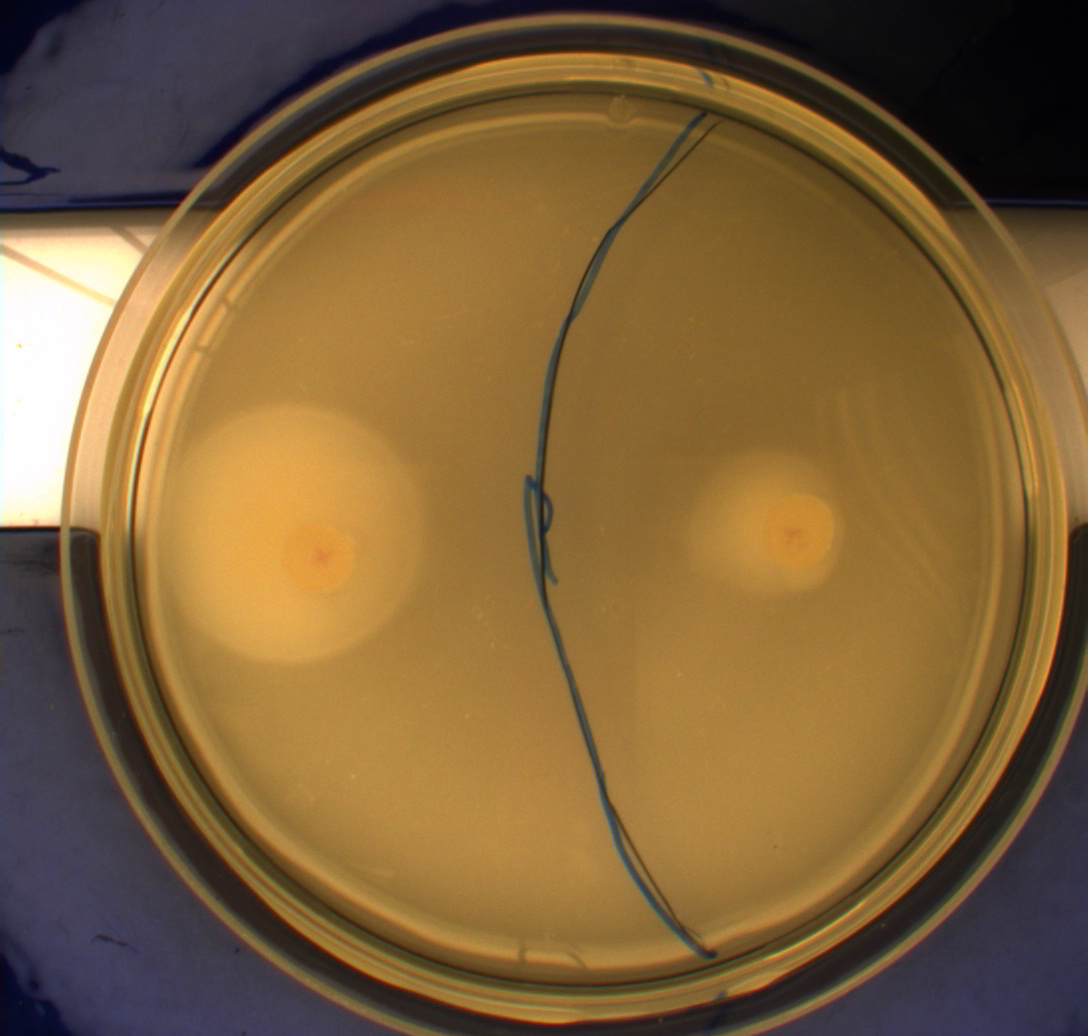
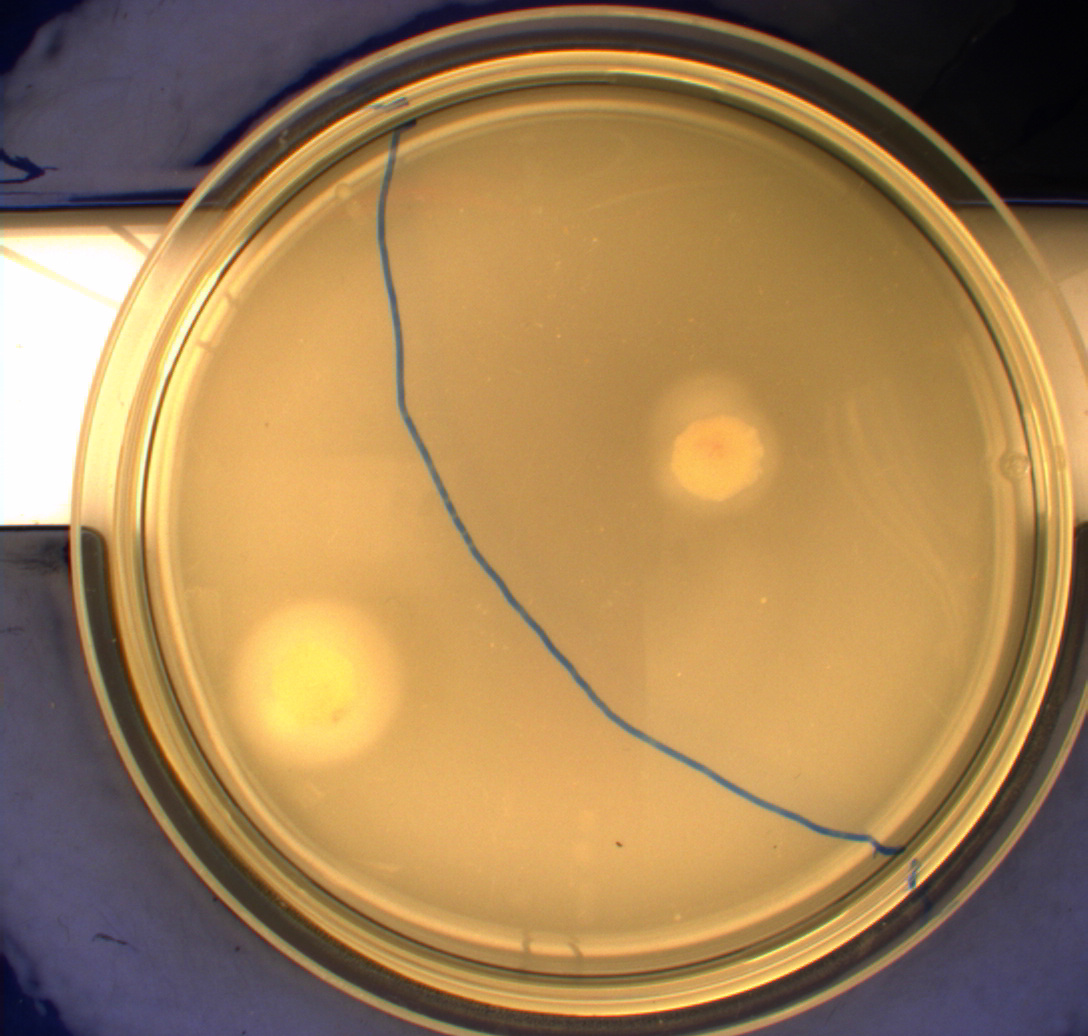
| - | In this experiment we shone blue light on one half of the plate | + | In this experiment, we shone blue light on one half of the plate while the other half was in the dark. We then placed one colony in each half and observed their motility pattern after 24 hours. We did this for three different strains of ''E. coli'': DH5alpha, Wildtype Mg1655 and Mg1655 containing a plasmid with our photosensor constitutively on. See the results for yourself: Light shone on the right half of the plates, the left half was in the dark.<br> |
[[Image:Team-SDU-Denmark-MG1655.JPG|250px|MG1655]] | [[Image:Team-SDU-Denmark-MG1655.JPG|250px|MG1655]] | ||
[[Image:Team-SDU-Denmark-Photosensor.JPG|250px|Photosensor]] | [[Image:Team-SDU-Denmark-Photosensor.JPG|250px|Photosensor]] | ||
[[Image:Team-SDU-Denmark-DH5alpha.JPG|250px|DH5alpha]]<br> | [[Image:Team-SDU-Denmark-DH5alpha.JPG|250px|DH5alpha]]<br> | ||
| - | Interpreting these results is not totally obvious since the effect of different motility patterns can be counter intuitive in semisolid agar. An explanation as to how different motility patterns show up on semisolid agar plates is explained by Englert et al. in the book "Microfluidic techniques for the analysis of bacterial chemotaxis":<br><br> | + | Interpreting these results is not totally obvious since the effect of different motility patterns can be counter intuitive in semisolid agar. An explanation as to how different motility patterns show up on semisolid agar plates is explained by Englert ''et al.'' in the book "Microfluidic techniques for the analysis of bacterial chemotaxis":<br><br> |
| - | ''Polymerized agar consists of chains of extended polymers permeated by water-filled channels. At low agar concentrations (0.25– 0.4% semisolid agar) the channels are sufficiently large that the bacteria can swim through them. In the absence of chemoeffectors, cells conduct a 3D random walk through the agar matrix much as they would in liquid and spread out randomly from the point of inoculation. Since the cells are typically growing, the result is an expanding colony that forms within the agar. This “pseudotaxis” occurs in the absence of any tactic behavior. Mutant | + | ''Polymerized agar consists of chains of extended polymers permeated by water-filled channels. At low agar concentrations (0.25– 0.4% semisolid agar) the channels are sufficiently large that the bacteria can swim through them. In the absence of chemoeffectors, cells conduct a 3D random walk through the agar matrix much as they would in liquid and spread out randomly from the point of inoculation. Since the cells are typically growing, the result is an expanding colony that forms within the agar. This “pseudotaxis” occurs in the absence of any tactic behavior. Mutant cells that only swim smoothly get trapped cul de sacs in the agar matrix (16), and their colonies do not expand significantly faster than those of nonmotile cells.'' <br><br> |
| - | cells that only swim smoothly get trapped cul de sacs in the agar matrix (16), and their colonies do not expand significantly faster than those of nonmotile cells.'' | + | |
| - | + | ||
| - | + | ||
| + | This means that if the photosensor leads to a reduced tumbling rate when exposed to blue light, it should look a lot like the colony of DH5alpha which was exposed to blue light. This is exactly the case which proves that the photosensor has an effect on the tumbling rate of the bacteria. The photosensor colony in the dark behaved like the Mg1655 colony in the dark and spread out. This is normal as cells that do not tend to either tumble or run a lot, will spread the most. <br><br> | ||
=== Video microscopy results === | === Video microscopy results === | ||
Revision as of 13:08, 20 October 2010
 "
"