Team:SDU-Denmark/project-r
From 2010.igem.org
(Difference between revisions)
(→Results) |
(→Results) |
||
| Line 10: | Line 10: | ||
=== Photosensor === | === Photosensor === | ||
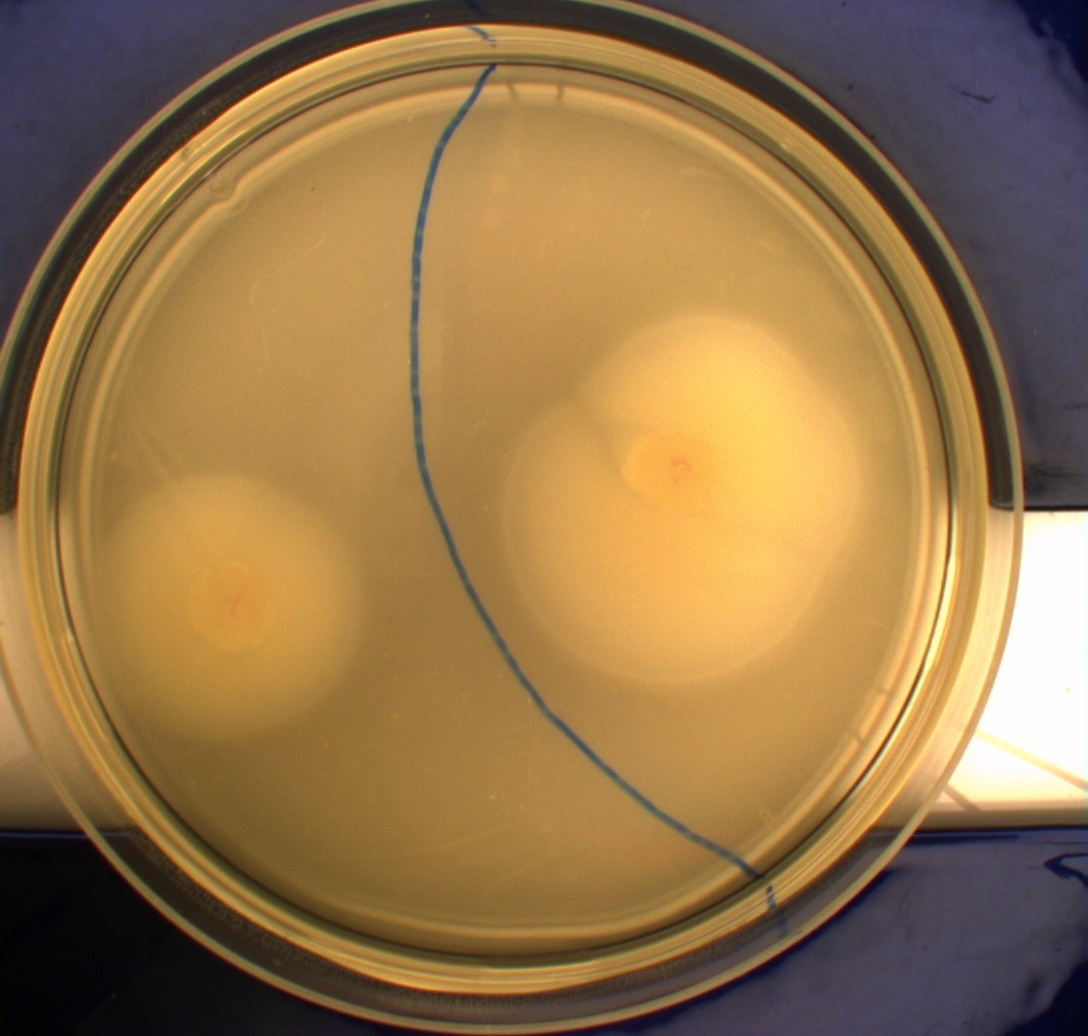
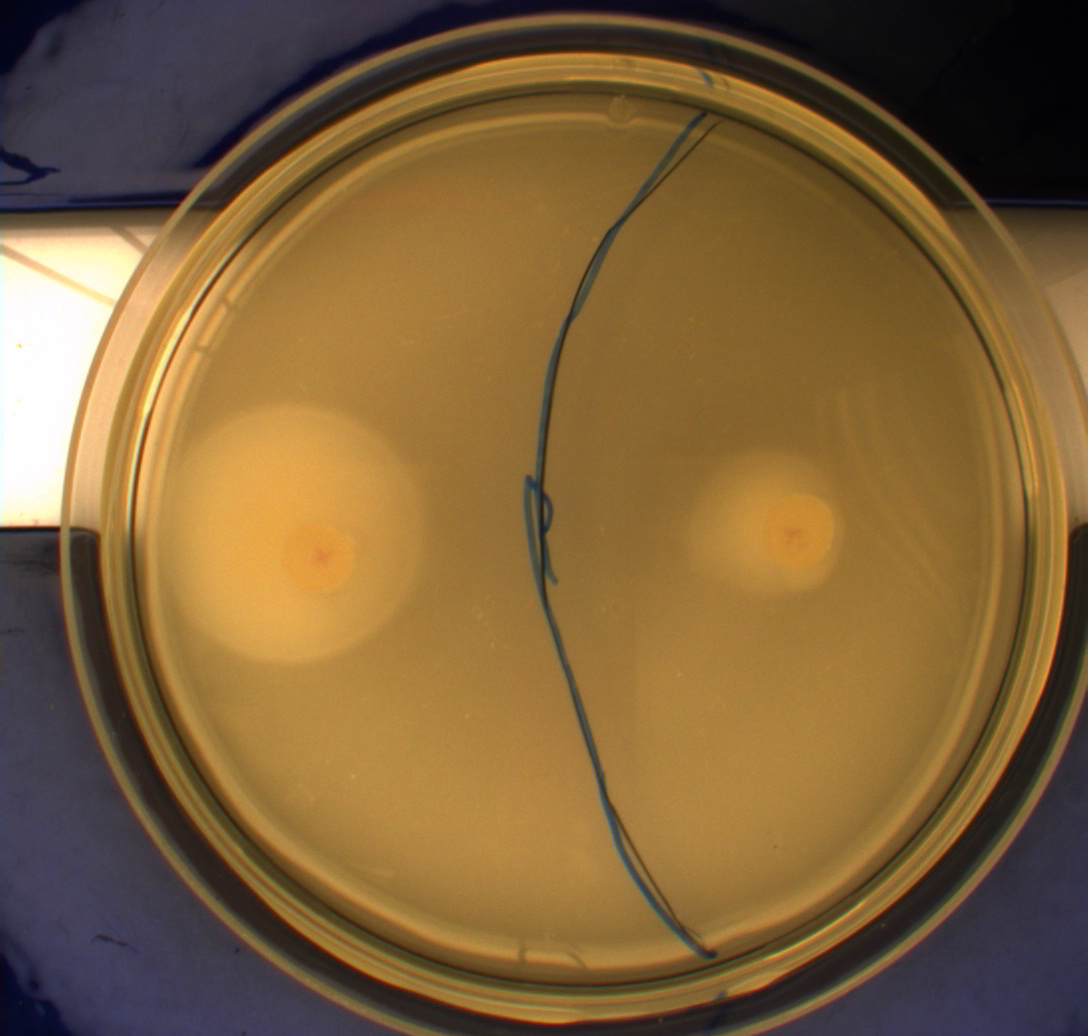
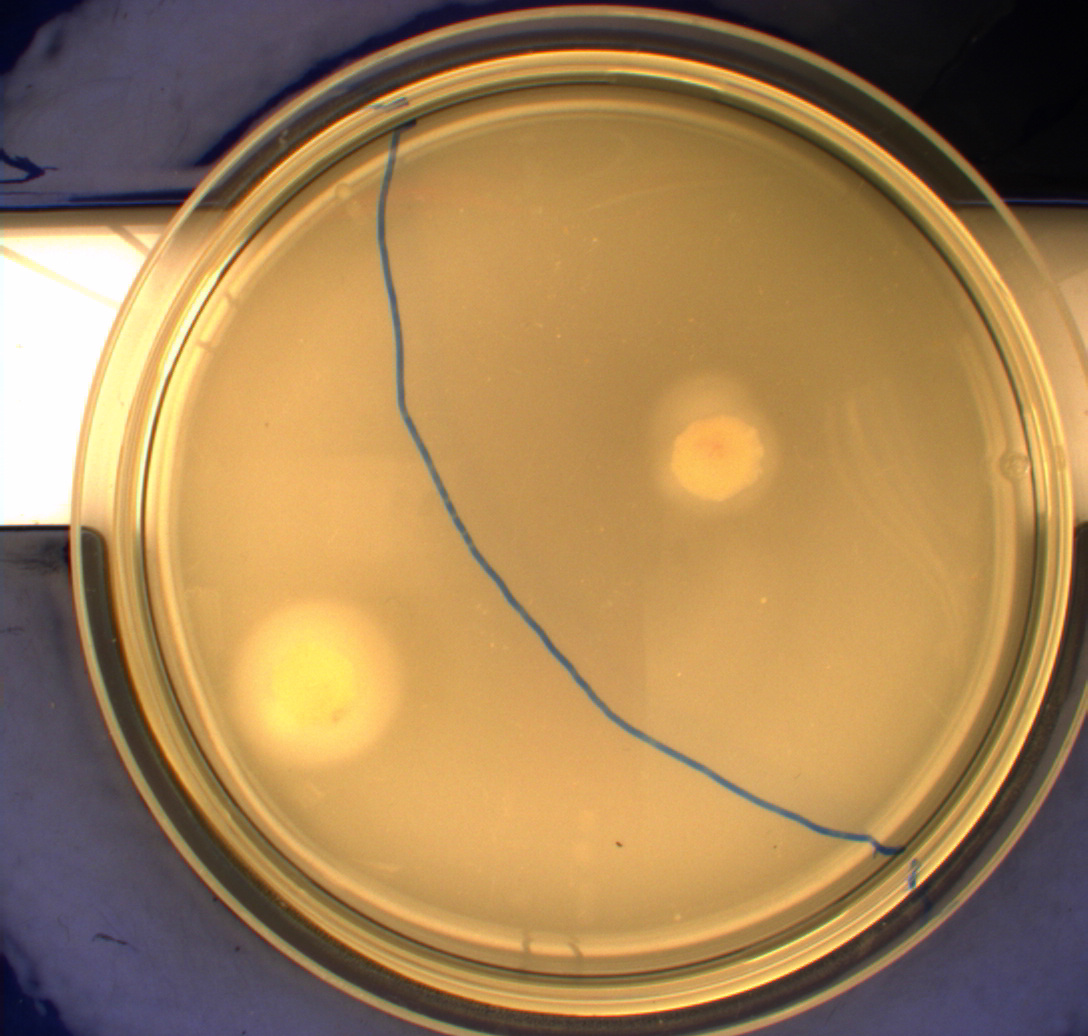
| - | '''Motility assay:''' In this experiment we shone blue light on one half of the plate, while the other half was in the dark. We then placed one colony in each half and observed their motility pattern after 24 hours. We did this for three different strains of E.Coli - DH5alpha, Wildtype Mg1655 and Mg1655 containing a plasmid with our photosensor constitutively on. See the results for yourself:<br> | + | '''Motility assay:'''<br> |
| + | In this experiment we shone blue light on one half of the plate, while the other half was in the dark. We then placed one colony in each half and observed their motility pattern after 24 hours. We did this for three different strains of E.Coli - DH5alpha, Wildtype Mg1655 and Mg1655 containing a plasmid with our photosensor constitutively on. See the results for yourself:<br> | ||
Light shone on the right half of the plates, the left half was in the dark.<br> | Light shone on the right half of the plates, the left half was in the dark.<br> | ||
[[Image:Team-SDU-Denmark-MG1655.JPG|250px|thumb|right|MG1655]] | [[Image:Team-SDU-Denmark-MG1655.JPG|250px|thumb|right|MG1655]] | ||
| Line 23: | Line 24: | ||
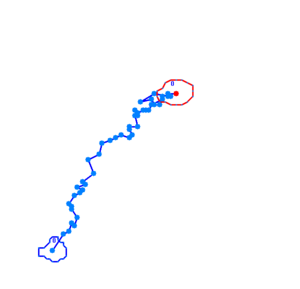
| - | '''Video microscopy results:''' | + | '''Video microscopy results:'''<br> |
Bacteria containing the photosensor will exhibit a lowered tumbling rate when exposed to blue light (wavelengths around 350nm - 450nm). This was analysed with the help of video microscopy and the open source software [http://db.cse.ohio-state.edu/CellTrack/ "CellTrack"]. The individual cells trajectory was tracked and their speed measured. The tracking results are as follows: | Bacteria containing the photosensor will exhibit a lowered tumbling rate when exposed to blue light (wavelengths around 350nm - 450nm). This was analysed with the help of video microscopy and the open source software [http://db.cse.ohio-state.edu/CellTrack/ "CellTrack"]. The individual cells trajectory was tracked and their speed measured. The tracking results are as follows: | ||
<br> | <br> | ||
| Line 35: | Line 36: | ||
=== Retinal === | === Retinal === | ||
| - | '''UV-Vis spectrophotometer determination of beta-carotene production: ''' In this experiment cells was prepare and harvested according to protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#EX1.1]. This experiment was preformed with four different strain of E. Coli – wildtype Top10, wildtype MG1655, Top10 containing PSB1A2 with K274210 constitutively active and MG1655 containing PSB1A2 with K274210 constitutively active. The resulting graphs is presented beneath the text<br> | + | '''UV-Vis spectrophotometer determination of beta-carotene production:'''<br> |
| + | In this experiment cells was prepare and harvested according to protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#EX1.1]. This experiment was preformed with four different strain of E. Coli – wildtype Top10, wildtype MG1655, Top10 containing PSB1A2 with K274210 constitutively active and MG1655 containing PSB1A2 with K274210 constitutively active. The resulting graphs is presented beneath the text<br> | ||
In order assess the results a standard dilution of beta-carotene: 1mM, 100 µM, 50 µM, 25 µM, 10 µM, 5 µM, 1 µM, 100 nM and 10 nM. The standart dilutions was also measured on the spectrophotometer. The resulting graphs is presented beneath the text<br> | In order assess the results a standard dilution of beta-carotene: 1mM, 100 µM, 50 µM, 25 µM, 10 µM, 5 µM, 1 µM, 100 nM and 10 nM. The standart dilutions was also measured on the spectrophotometer. The resulting graphs is presented beneath the text<br> | ||
Revision as of 10:25, 18 October 2010
 "
"