Team:SDU-Denmark/project-p
From 2010.igem.org
(Difference between revisions)
(→Characterization of parts) |
(→K343007) |
||
| Line 25: | Line 25: | ||
After this we could conclude that the part had an impact on the bacterial motility, but we had to find out which. This lead us to our next experiment, which was intended for determining what happened to the tumbling frequency:<br> | After this we could conclude that the part had an impact on the bacterial motility, but we had to find out which. This lead us to our next experiment, which was intended for determining what happened to the tumbling frequency:<br> | ||
<br> | <br> | ||
| - | ''' | + | '''3. Videomicroscopy and computer analysis:'''<br> |
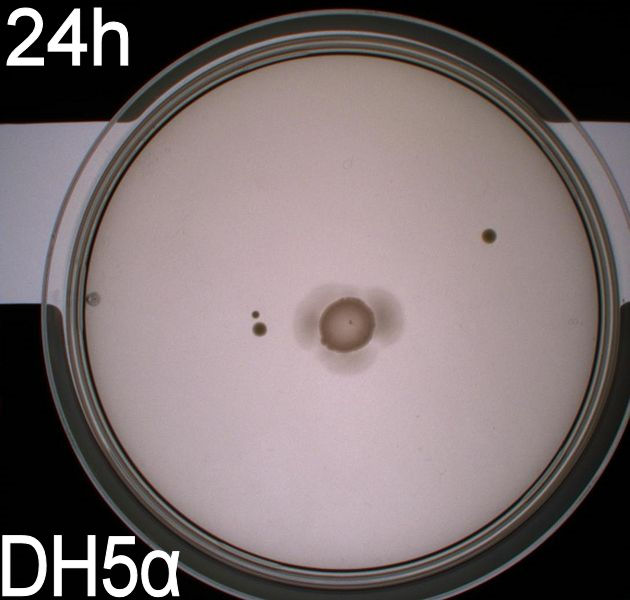
To get an exact idea of what is happening to the bacteria when adding the photosensor we decided on doing video microscopy of motile bacteria. The bacterial cultures for this analysis were incubated at 37° C, 180 RPM overnight, afterwards the optical density at 550nm was measured and the cultures were diluted to an OD550 of 0,02. They were then again incubated at 37 degrees and 180 RPM until their optical density reached 0,5. At this point we added retinal to the culture at a concentration of 5 uM and incubated the cells in the same conditions for another two hours. On top of phototactic bacteria we cultivated MG1655 (wildtype) and DH5alpha (non-motile) as controls. These were grown until an OD550 of 0,5 with the same methods as described for the modified culture, at this point these strains were ready for microscopy. <br> | To get an exact idea of what is happening to the bacteria when adding the photosensor we decided on doing video microscopy of motile bacteria. The bacterial cultures for this analysis were incubated at 37° C, 180 RPM overnight, afterwards the optical density at 550nm was measured and the cultures were diluted to an OD550 of 0,02. They were then again incubated at 37 degrees and 180 RPM until their optical density reached 0,5. At this point we added retinal to the culture at a concentration of 5 uM and incubated the cells in the same conditions for another two hours. On top of phototactic bacteria we cultivated MG1655 (wildtype) and DH5alpha (non-motile) as controls. These were grown until an OD550 of 0,5 with the same methods as described for the modified culture, at this point these strains were ready for microscopy. <br> | ||
The actual microscopy was done on a (insert name) microscope with an optical magnification of 1000x. <br> | The actual microscopy was done on a (insert name) microscope with an optical magnification of 1000x. <br> | ||
| Line 40: | Line 40: | ||
<object width="320"><param name="movie" value="http://www.youtube.com/v/HEKbYjyWNJc?hl=de&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/HEKbYjyWNJc?hl=de&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="300"></embed></object> <object width="320"><param name="movie" value="http://www.youtube.com/v/bdg36HRHGRc?hl=de&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/bdg36HRHGRc?hl=de&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="320" ></embed></object> </html><br><br> | <object width="320"><param name="movie" value="http://www.youtube.com/v/HEKbYjyWNJc?hl=de&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/HEKbYjyWNJc?hl=de&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="300"></embed></object> <object width="320"><param name="movie" value="http://www.youtube.com/v/bdg36HRHGRc?hl=de&fs=1"></param><param name="allowFullScreen" value="true"></param><param name="allowscriptaccess" value="always"></param><embed src="http://www.youtube.com/v/bdg36HRHGRc?hl=de&fs=1" type="application/x-shockwave-flash" allowscriptaccess="always" allowfullscreen="true" width="320" ></embed></object> </html><br><br> | ||
When played at real time speed these videos seem slow and not showing any tendency, though when played at 4 times the normal speed, the photosensor will seem to show slightly increased motility when exposed to blue light. Even though the bacteria displayed disappointing motility, we went on and did computer analysis on the videos by the help of the open source software "CellTrack" (Link!). The paths that were mapped showed a longer distance traveling for the photosensor exposed to blue light, when compared with the rest of the samples. This could indicated a decreased tumbling frequency, but since the bacteria's motility was very low, the results were stamped as unreliable and a new protocol had to be devised.<br><br> | When played at real time speed these videos seem slow and not showing any tendency, though when played at 4 times the normal speed, the photosensor will seem to show slightly increased motility when exposed to blue light. Even though the bacteria displayed disappointing motility, we went on and did computer analysis on the videos by the help of the open source software "CellTrack" (Link!). The paths that were mapped showed a longer distance traveling for the photosensor exposed to blue light, when compared with the rest of the samples. This could indicated a decreased tumbling frequency, but since the bacteria's motility was very low, the results were stamped as unreliable and a new protocol had to be devised.<br><br> | ||
| - | '''Computerized analysis of the bacterial motility with the Thor prototype by Unisensor A/S and the Unify software:'''<br> | + | '''4. Computerized analysis of the bacterial motility with the Thor prototype by Unisensor A/S and the Unify software:'''<br> |
For this experiment we changed our protocol for cultivating swimming bacteria.<br> | For this experiment we changed our protocol for cultivating swimming bacteria.<br> | ||
After a series of experiments we settled on a pipette tip from a freezing culture inoculated in 5 ml LB media. The culture was then incubated for 12 hours at 22° C and 160 RPM. Afterwards the unmodified cultures were diluted 1:10 in Motility buffer (reference Adler), at which point thez were ready for microscopy. At this point retinal is added to the strain containing the photosensor and it continues to grow for another two hours in darkness, afterwards they were also diluted in motility buffer.<br> | After a series of experiments we settled on a pipette tip from a freezing culture inoculated in 5 ml LB media. The culture was then incubated for 12 hours at 22° C and 160 RPM. Afterwards the unmodified cultures were diluted 1:10 in Motility buffer (reference Adler), at which point thez were ready for microscopy. At this point retinal is added to the strain containing the photosensor and it continues to grow for another two hours in darkness, afterwards they were also diluted in motility buffer.<br> | ||
Revision as of 13:46, 24 October 2010
 "
"