Team:SDU-Denmark/labnotes7
From 2010.igem.org
(Difference between revisions)
(→Lab notes (23/9 - 29/9)) |
(→Restriction Digest) |
||
| (10 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<div id="leftcolumn"> | <div id="leftcolumn"> | ||
| - | = Lab notes (23/ | + | = Lab notes (23/8 - 29/8) = |
__TOC__ | __TOC__ | ||
| + | |||
| + | == Photosensor == | ||
| + | === Insertion of B0015 in pSB3C5 and pSB3T5 === | ||
| + | |||
| + | '''Date:''' 8/23 - 8/29 2010<Br> | ||
| + | '''Done By:''' Maria and Lc<Br> | ||
| + | '''Protocol:''' [https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1 CP1.1][https://2010.igem.org/Team:SDU-Denmark/protocols#DE1.3 DE1.3][https://2010.igem.org/Team:SDU-Denmark/protocols#RD1.1 RD1.1][https://2010.igem.org/Team:SDU-Denmark/protocols#LG1.2 LG1.2][https://2010.igem.org/Team:SDU-Denmark/protocols#CC1.1 CC1.1][https://2010.igem.org/Team:SDU-Denmark/protocols#TR1.1 TR1.1][https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.3 CP1.3]<Br> | ||
| + | ==== Pfu PCR amplification and purification of B0015 ==== | ||
| + | '''Date:''' 8/23 2010<Br> | ||
| + | '''Done By:''' Maria and Lc<Br> | ||
| + | '''Protocol:'''[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1 CP1.1]<br> | ||
| + | '''Notes:'''<br> | ||
| + | 4 PCR reactions are prepared. 2uL Miniprep of pSB1AK3 (white 43) are distrubuted in each tube. PCR tubes are marked B00153.A-D.<br> | ||
| + | Premix x5:<br> | ||
| + | <table style="text-align: left;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td>pfu buffer + MgSO<small>4</small></td> | ||
| + | <td>25uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>dNTP mix</td> | ||
| + | <td>7.5uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>VF2 primer</td> | ||
| + | <td>7.5uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>VR primer</td> | ||
| + | <td>7.5uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>H<small>2</small>0</td> | ||
| + | <td>190uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pfu polymerase </td> | ||
| + | <td>2uL</td> | ||
| + | </tr> | ||
| + | </table><br><br> | ||
| + | PCR program:<br> | ||
| + | <table style="text-align: left;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td>start</td> | ||
| + | <td>94C</td> | ||
| + | <td>3min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>denaturating</td> | ||
| + | <td>94C</td> | ||
| + | <td>2min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>annealing</td> | ||
| + | <td>55C</td> | ||
| + | <td>30s</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>elongation</td> | ||
| + | <td>72C</td> | ||
| + | <td>30s</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>go to</td> | ||
| + | <td>2</td> | ||
| + | <td>rep.29x</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>end</td> | ||
| + | <td>72C</td> | ||
| + | <td>5min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>hold</td> | ||
| + | <td>4C</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table><br> | ||
| + | 5uL of the PCR product was loaded onto a 2% agarose gel. Gene ruler 100bp DNA ladder was used ad marker.<br><br> | ||
| + | '''Results:'''<br> | ||
| + | '''Analysis:'''<br> | ||
| + | A strong band was observed at app. 450bp, and the rest of the PCR product was purified according to protocol using the GFX purification kit.<br> | ||
| + | End conc.: 48ng/uL <br><br> | ||
| + | |||
| + | ==== Restriction digest of B0015 ==== | ||
| + | '''Date:''' 8/24 2010<Br> | ||
| + | '''Done By:''' Maria and Lc<Br> | ||
| + | '''Protocol:'''[https://2010.igem.org/Team:SDU-Denmark/protocols#RD1.1 RD1.1][https://2010.igem.org/Team:SDU-Denmark/protocols#DE1.3 DE1.3]<br> | ||
| + | '''Notes:'''<br> | ||
| + | Restriction mixture B0015:<br> | ||
| + | <table style="text-align: left;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td>H<small>2</small>O</td> | ||
| + | <td>38uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>FD green buffer</td> | ||
| + | <td>8uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>EcoRI</td> | ||
| + | <td>4uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>PstI</td> | ||
| + | <td>4uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>B0015</td> | ||
| + | <td>30uL</td> | ||
| + | </tr> | ||
| + | </table><br><br> | ||
| + | |||
| + | The digested sample was loaded onto a 2% agarose extraction gel. Uncut B0015 was used as controle. Gene ruler 100bp DNA ladder was used as marker.<br> | ||
| + | DNA was extracted from gel according to protocol.<br><br> | ||
| + | '''Results:'''<br> | ||
| + | DNA conc: | ||
| + | <table style="text-align: left;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td>sample</td> | ||
| + | <td>conc. (ng/uL)</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>B0015</td> | ||
| + | <td>6.5</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB3C5</td> | ||
| + | <td>50.8</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB3T5</td> | ||
| + | <td>9.9</td> | ||
| + | </tr> | ||
| + | </table><br><br> | ||
| + | '''Analysis:''' | ||
| + | the purified DNA was used for ligation.<br><br> | ||
| + | |||
| + | ==== Ligation of PS and pSB1C3 and pCB1AK3 ==== | ||
| + | '''Date:''' 8/24 2010<Br> | ||
| + | '''Done By:''' Maria and Lc<Br> | ||
| + | '''Protocol:'''[https://2010.igem.org/Team:SDU-Denmark/protocols#LG1.2 LG1.2]<br> | ||
| + | '''Notes:'''<br> | ||
| + | For each of the ligations three ligation mixtures were prepared. vector concentrations of 10n0g/uL (pSB3C5) and 50ng/uL (pSB3T5) respectively was used for each mixture. Appropiate amount of insert was added to reach vector:insert ratios of 1:1, 1:3 and 1:6 respectively. <br> | ||
| + | Ligation mixtures (B0015 in pSB3C5):<br> | ||
| + | <table style="text-align: left;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>L1</td> | ||
| + | <td>L2</td> | ||
| + | <td>L3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>T4 ligase buffer</td> | ||
| + | <td>2uL</td> | ||
| + | <td>2uL</td> | ||
| + | <td>2uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>T4 ligase</td> | ||
| + | <td>1uL</td> | ||
| + | <td>1uL</td> | ||
| + | <td>1uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB3C5</td> | ||
| + | <td>2uL</td> | ||
| + | <td>2uL</td> | ||
| + | <td>2uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>B0015 </td> | ||
| + | <td>0.7uL</td> | ||
| + | <td>2uL</td> | ||
| + | <td>4.5uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>H<small>2</small>0</td> | ||
| + | <td>14.3uL</td> | ||
| + | <td>13uL</td> | ||
| + | <td>10.5uL</td> | ||
| + | </tr> | ||
| + | </table><br> | ||
| + | Ligation mixtures (B0015 in pSB3T5):<br> | ||
| + | <table style="text-align: left;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td></td> | ||
| + | <td>L1</td> | ||
| + | <td>L2</td> | ||
| + | <td>L3</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>T4 ligase buffer</td> | ||
| + | <td>2uL</td> | ||
| + | <td>2uL</td> | ||
| + | <td>2uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>T4 ligase</td> | ||
| + | <td>1uL</td> | ||
| + | <td>1uL</td> | ||
| + | <td>1uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>pSB3T5</td> | ||
| + | <td>5uL</td> | ||
| + | <td>5uL</td> | ||
| + | <td>5uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>B0015 </td> | ||
| + | <td>0.5uL</td> | ||
| + | <td>1uL</td> | ||
| + | <td>2uL</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>H<small>2</small>0</td> | ||
| + | <td>11.5uL</td> | ||
| + | <td>11uL</td> | ||
| + | <td>10uL</td> | ||
| + | </tr> | ||
| + | </table><br><br> | ||
| + | The samples was incubated at 17C ON at used for transformation<br><br> | ||
| + | |||
| + | ==== Transfomation of ligated plasmid in Top 10 E.coli ==== | ||
| + | |||
| + | '''Date:''' 8/25 2010<Br> | ||
| + | '''Done By:''' Maria and Lc<Br> | ||
| + | '''Protocol:'''[https://2010.igem.org/Team:SDU-Denmark/protocols#CC1.1 CC1.1][https://2010.igem.org/Team:SDU-Denmark/protocols#TR1.1 TR1.1]<br> | ||
| + | '''Notes:'''<br> | ||
| + | The compotent cells and transformation was carried out according to protocol.<br><br> | ||
| + | '''Results:'''<br> | ||
| + | the controle plates were okay, and there were many colonies on plates with cells transformed with either of the ligation mixtures..<br><br> | ||
| + | '''Analysis:'''<br> | ||
| + | The transformation was successfull and colonies was selected and used in coloni PCR.<br> | ||
| + | --[[User:Tipi|Tipi]] 13:41, 26 September 2010 (UTC)<br><br> | ||
| + | === Colony PCR on B0017 === | ||
| + | Date: 27/8<br> | ||
| + | Done by: LC<br> | ||
| + | Methods: PCR<br> | ||
| + | Protocols: [https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.3 CP1.3]<br> | ||
| + | Notes: <br> | ||
| + | Premix:<br> | ||
| + | 12,5 µl 10xTAQ Buffer<br> | ||
| + | 5 µl MgCl2 <br> | ||
| + | 5 µl VF2 <br> | ||
| + | 5 µl VR <br> | ||
| + | 2,5 µl dNTP<br> | ||
| + | 17,5 µl H2O<br> | ||
| + | 5/8 µl TAQ Polymerase<br> | ||
| + | <br> | ||
| + | 9,5 µl Premix were added to 15 µl of H2O containing the lysed cells.<br> | ||
| + | <br> | ||
| + | PCR Program:<br> | ||
| + | <table style="text-align: left;" border="1" cellpadding="2" | ||
| + | cellspacing="2"> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Start<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">94 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">2 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Denaturing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">94 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">1 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Annealing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">55 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">1 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Elongation<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">72 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">30 sec<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Goto2<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">rep<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">29x<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">End<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">72 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">3 min<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Hold<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">4 C<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;"><br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </table> | ||
| + | <br> | ||
| + | <br> | ||
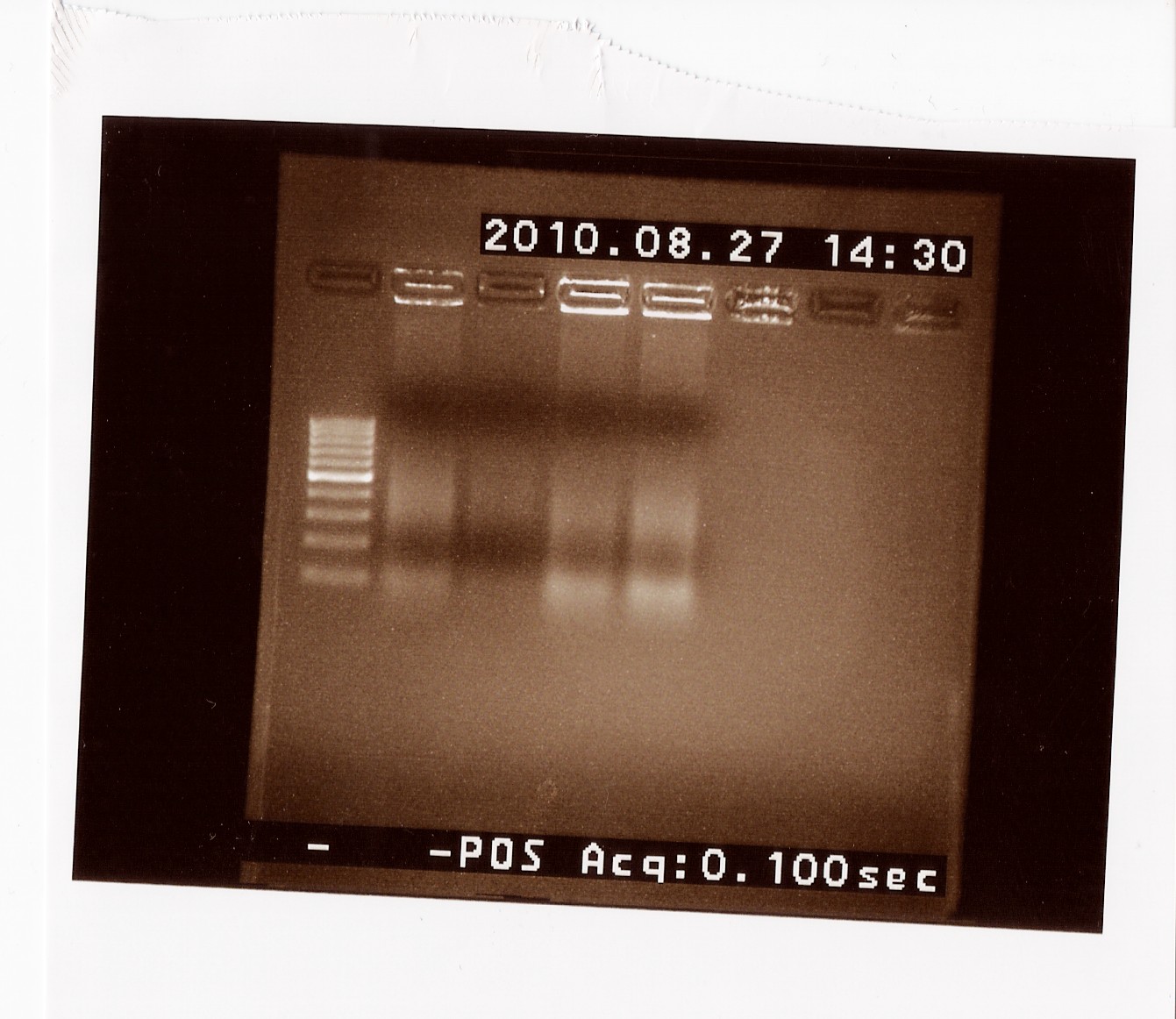
| + | Results: <br> | ||
| + | [[Image:Team-SDU-Denmark-cPCRB0017.jpg|300px]] <br> | ||
| + | No useable results, only unclear bands around 120 BP, which seem to be the result of mispriming with VR on B0010. | ||
| + | |||
| + | == Retinal == | ||
==== Futher PCR on POT2 with NinaB (New Primers NO. 5) ==== | ==== Futher PCR on POT2 with NinaB (New Primers NO. 5) ==== | ||
| Line 12: | Line 338: | ||
Done by: Marie & Tommy<br> | Done by: Marie & Tommy<br> | ||
Methods: PCR<br> | Methods: PCR<br> | ||
| - | protocos: | + | protocos:[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1 CP1.1] |
Notes:<br> | Notes:<br> | ||
NinB2fw and NinaB2rv was used.<br> | NinB2fw and NinaB2rv was used.<br> | ||
| Line 23: | Line 349: | ||
</head> | </head> | ||
<body> | <body> | ||
| - | <table style="text-align: left; width: | + | <table style="text-align: left; width: 300px;" border="1" |
cellpadding="2" cellspacing="2"> | cellpadding="2" cellspacing="2"> | ||
<tr> | <tr> | ||
| Line 71: | Line 397: | ||
Start date: 24/8<br> | Start date: 24/8<br> | ||
Methods: Ligation, Competent cells, Transformation<br> | Methods: Ligation, Competent cells, Transformation<br> | ||
| - | Protocols: | + | Protocols: [https://2010.igem.org/Team:SDU-Denmark/protocols#LG1.1 LG1.1], [https://2010.igem.org/Team:SDU-Denmark/protocols#CC1.1 CC1.1], [https://2010.igem.org/Team:SDU-Denmark/protocols#TR1.1 TR1.1] |
==== DNA purification from PCR ==== | ==== DNA purification from PCR ==== | ||
| Line 97: | Line 423: | ||
</head> | </head> | ||
<body> | <body> | ||
| - | <table style="text-align: left; width: | + | <table style="text-align: left; width: 100px;" border="1" |
cellpadding="2" cellspacing="2"> | cellpadding="2" cellspacing="2"> | ||
<tr> | <tr> | ||
| Line 137: | Line 463: | ||
Done by: Marie & Tommy<br> | Done by: Marie & Tommy<br> | ||
Methods: Ligation<br> | Methods: Ligation<br> | ||
| - | protocos: | + | protocos:[https://2010.igem.org/Team:SDU-Denmark/protocols#LG1.3 L1.3] |
Notes:<br> | Notes:<br> | ||
3 ligatons mixtures was made:<br> | 3 ligatons mixtures was made:<br> | ||
| Line 148: | Line 474: | ||
Done by: Marie & Tommy<br> | Done by: Marie & Tommy<br> | ||
Methods: Colony PCR<br> | Methods: Colony PCR<br> | ||
| - | protocos: | + | protocos:[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1 CP1.1] |
Notes:<br> | Notes:<br> | ||
15 colonies were picked form different plates, with differnt plasmid to insert ratio: colonie's 1,2,3,13 were picked from plates with 1:1 plasmid to insert ratio, colonie's 4,5,6 were picked from plates with 1:3 plasmid to insert ratio and colonie's 7,8,9,10,11,12,14,15 were picked from plates with 1:6 plasmid to insert ratio.<br> | 15 colonies were picked form different plates, with differnt plasmid to insert ratio: colonie's 1,2,3,13 were picked from plates with 1:1 plasmid to insert ratio, colonie's 4,5,6 were picked from plates with 1:3 plasmid to insert ratio and colonie's 7,8,9,10,11,12,14,15 were picked from plates with 1:6 plasmid to insert ratio.<br> | ||
| Line 159: | Line 485: | ||
</head> | </head> | ||
<body> | <body> | ||
| - | <table style="text-align: left; width: | + | <table style="text-align: left; width: 300px;" border="1" |
cellpadding="2" cellspacing="2"> | cellpadding="2" cellspacing="2"> | ||
<tr> | <tr> | ||
| Line 207: | Line 533: | ||
Done by: Marie & Tommy<br> | Done by: Marie & Tommy<br> | ||
Methods: Restriction Digest<br> | Methods: Restriction Digest<br> | ||
| - | protocos: | + | protocos:[https://2010.igem.org/Team:SDU-Denmark/protocols#RD1.1 RD1.1] |
Notes:<br> | Notes:<br> | ||
Restriction digest was performed on tubes 5,6,10 and 11 to test for insertion of ninaB (Correct orientation) XbaI and SPEI was used and a gel was run:<br> | Restriction digest was performed on tubes 5,6,10 and 11 to test for insertion of ninaB (Correct orientation) XbaI and SPEI was used and a gel was run:<br> | ||
| Line 215: | Line 541: | ||
Done by: Marie & Tommy<br> | Done by: Marie & Tommy<br> | ||
Methods: Colony PCR<br> | Methods: Colony PCR<br> | ||
| - | protocos: | + | protocos:[https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1 CP1.1] |
Notes:<br> | Notes:<br> | ||
8 new colonies (16-23) were chosen in addition to colonies 5,6,10 and 11 form 25I8-10 colonie PCR.<br> | 8 new colonies (16-23) were chosen in addition to colonies 5,6,10 and 11 form 25I8-10 colonie PCR.<br> | ||
| Line 226: | Line 552: | ||
</head> | </head> | ||
<body> | <body> | ||
| - | <table style="text-align: left; width: | + | <table style="text-align: left; width: 300px;" border="1" |
cellpadding="2" cellspacing="2"> | cellpadding="2" cellspacing="2"> | ||
<tr> | <tr> | ||
| Line 280: | Line 606: | ||
</head> | </head> | ||
<body> | <body> | ||
| - | <table style="text-align: left; width: | + | <table style="text-align: left; width: 300px;" border="1" |
cellpadding="2" cellspacing="2"> | cellpadding="2" cellspacing="2"> | ||
<tr> | <tr> | ||
Latest revision as of 22:31, 23 October 2010
 "
"