Team:SDU-Denmark/labnotes4
From 2010.igem.org
(Difference between revisions)
(→Checking if Pst1 site in FlhDC mut is not pressent vol.2) |
(→vol. 3) |
||
| Line 26: | Line 26: | ||
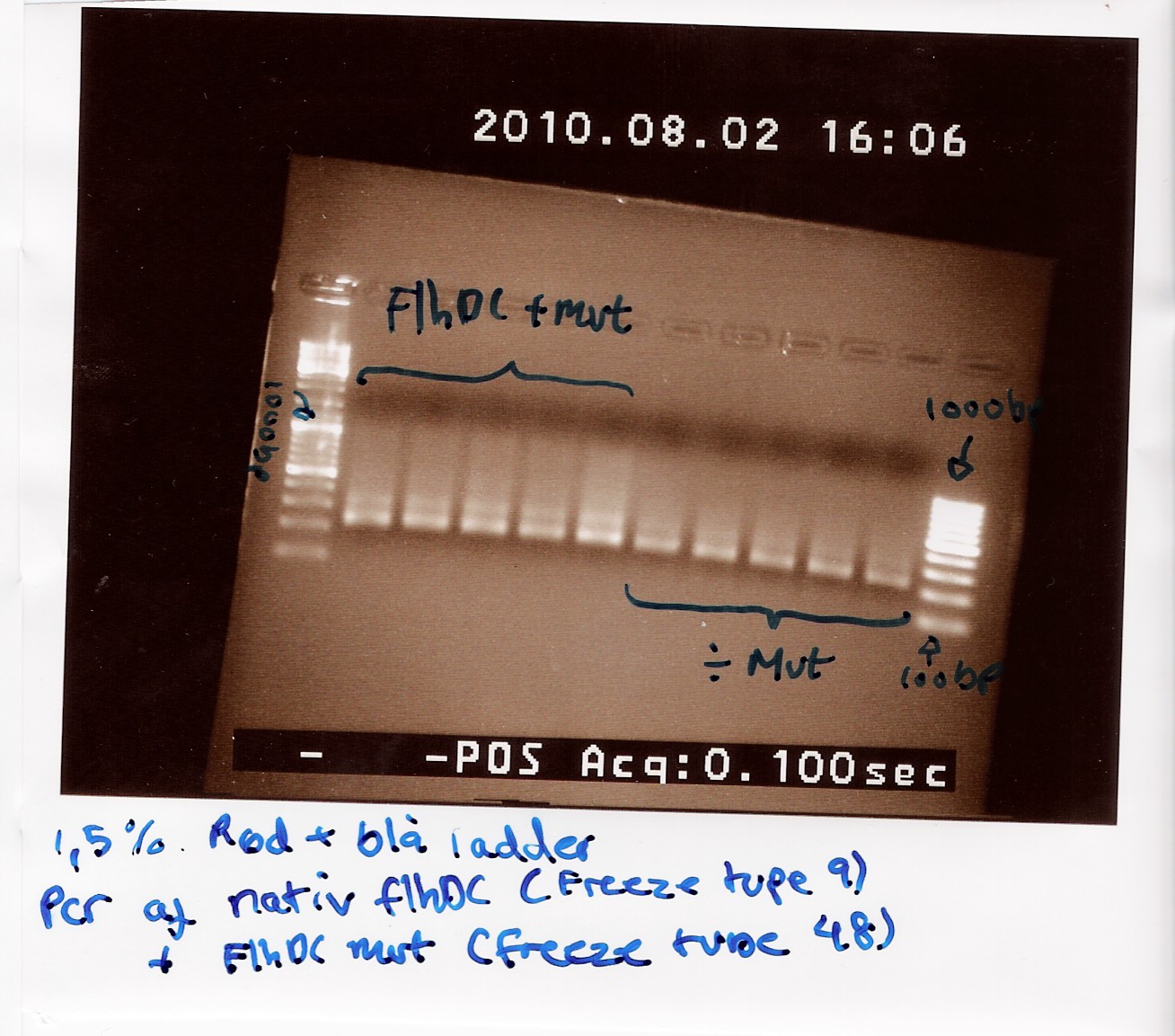
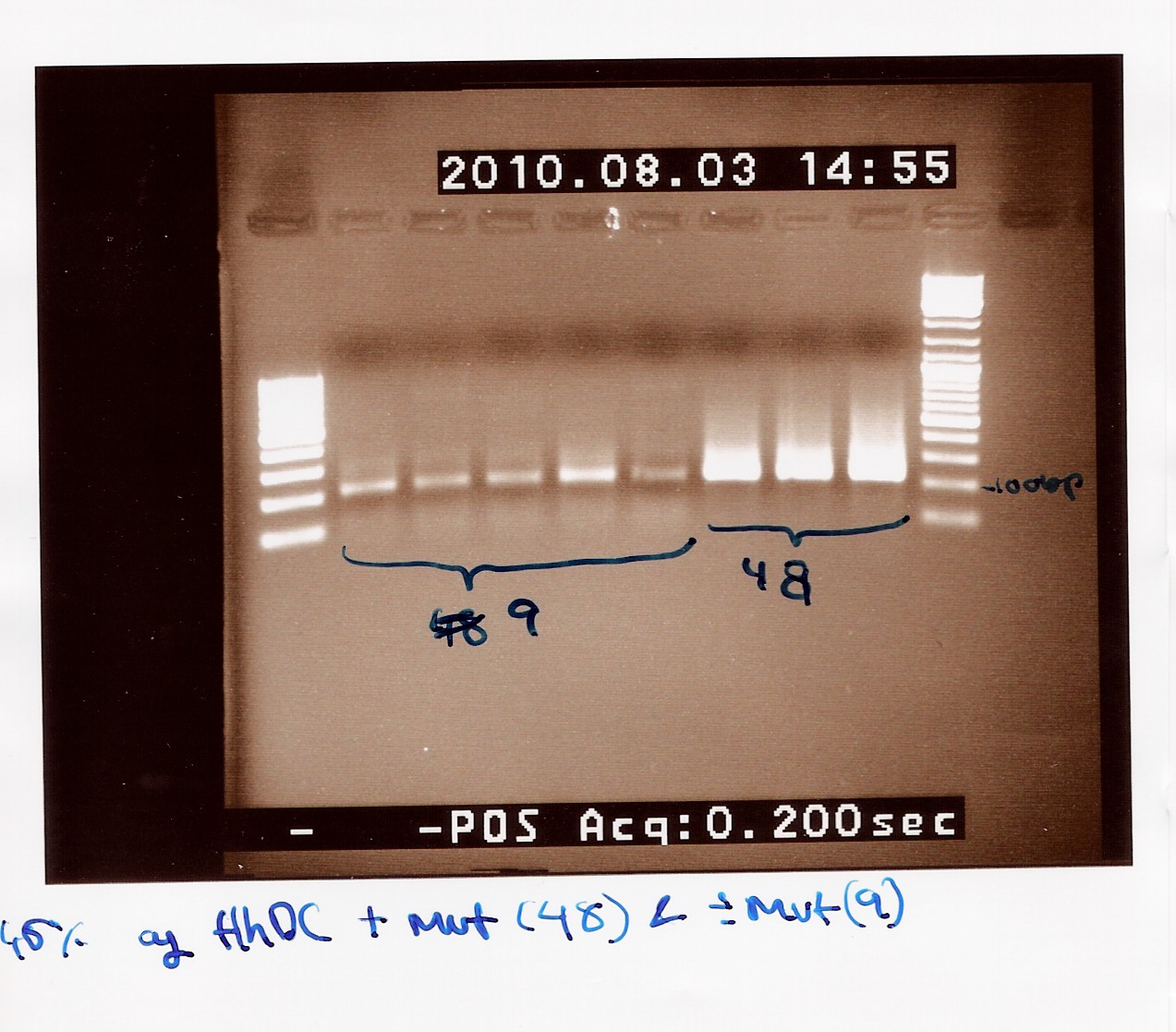
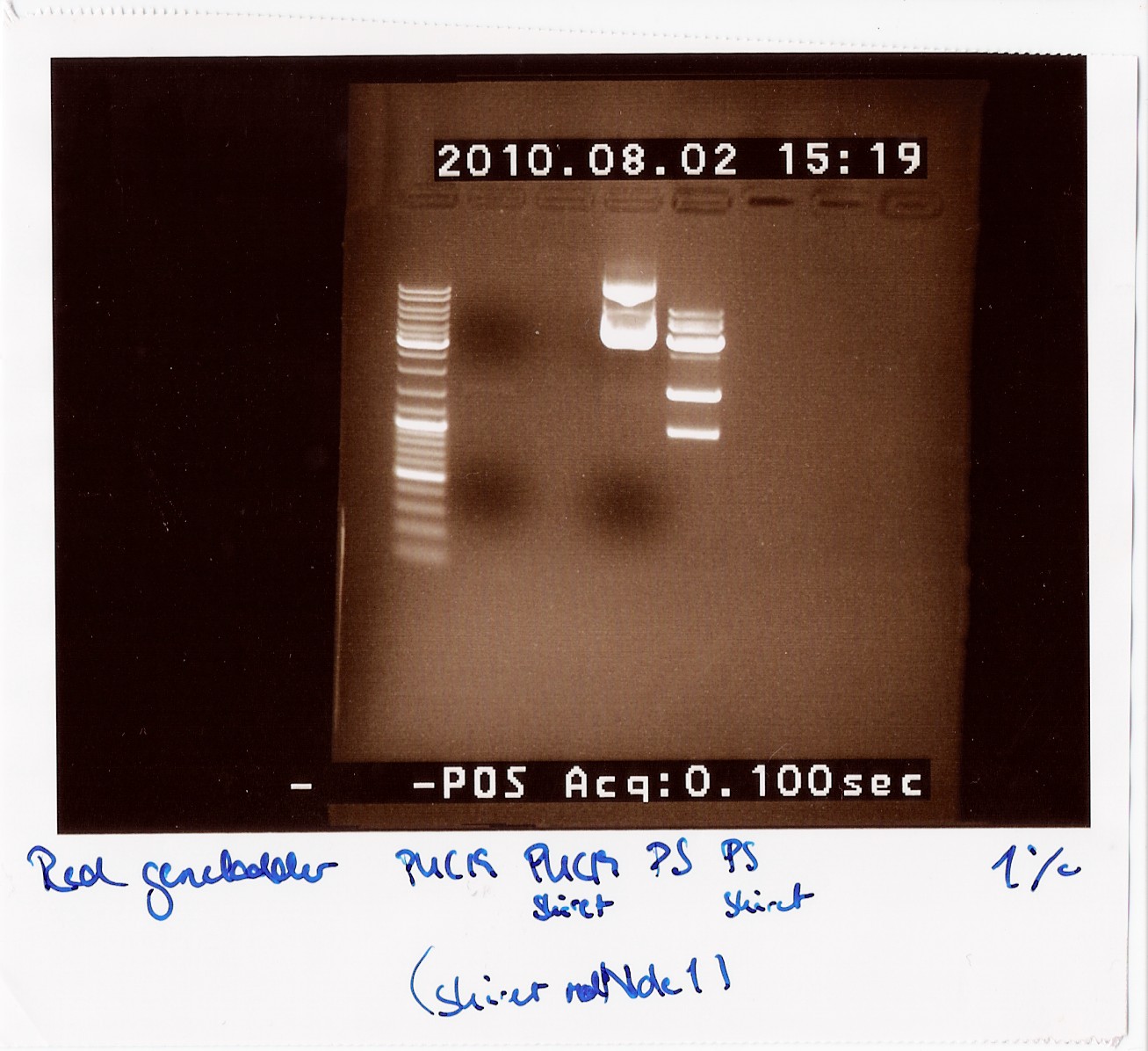
==== vol. 3 ==== | ==== vol. 3 ==== | ||
''Done by:'' Sheila & Louise <br> | ''Done by:'' Sheila & Louise <br> | ||
| - | ''Date:'' August | + | ''Date:'' August 3th <br> |
''Protocol:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#RD1.1 RD1.1] <br> | ''Protocol:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#RD1.1 RD1.1] <br> | ||
''Notes:'' The previous experiment was redone with the new primers. <br> Five samples of native FlhDC (FT 9) and 3 samples of FlhDC mut (FT 48) was loaded on a 1.5% gel with two ladders (100-1000 and 100-10000). <br> | ''Notes:'' The previous experiment was redone with the new primers. <br> Five samples of native FlhDC (FT 9) and 3 samples of FlhDC mut (FT 48) was loaded on a 1.5% gel with two ladders (100-1000 and 100-10000). <br> | ||
| Line 33: | Line 33: | ||
<br> | <br> | ||
--[[User:Louch07|Louch07]] 10:20, 5 August 2010 (UTC) | --[[User:Louch07|Louch07]] 10:20, 5 August 2010 (UTC) | ||
| + | |||
| + | === Colony PCR of FlhDCmut and DT (B0015) === | ||
| + | ''Done by:'' Maria, Pernille & Louise <br> | ||
| + | ''Date:'' August 4th <br> | ||
| + | ''Protocol:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.3 CP1.3] <br> | ||
| + | ''Notes:'' The FlhDC mut gene was previously incearted into two different plasmids (pSB3k3 and pSB1c3) and the DT was incearted into pSB3c5. These were grown and 8 colonies from different plates were picked<br><br> | ||
| + | FlhDCmut in pSB3k3, PCR-tube 1a-h <br> | ||
| + | - 1a, -b, -c were picked from plate L1.1b (30.07.10) <br> | ||
| + | - 1d, -e, -f were picked from plate L2.1a (30.07.10) <br> | ||
| + | - 1g, -h were picked from plate L3.2a (30.07.10) <br><br> | ||
| + | FlhDCmut in pSB1c3, PCR-tube 2a-h <br> | ||
| + | - 2a, -b were picked from plate L1.3b (30.07.10) <br> | ||
| + | - 2c, -d, -e were picked from plate L3.2b (30.07.10) <br> | ||
| + | - 2f, -g, -h were picked from plate L2.1b (30.07.10) <br><br> | ||
| + | DT(B0015) in pSB3c5, PCR-tube 3a-h* <br> | ||
| + | - 3a, -b were picked from plate L1.2a (30.07.10) <br> | ||
| + | - 3c, -d were picked from plate L2.1c (30.07.10) <br> | ||
| + | - 3e, -f were picked from plate L2.1b (30.07.10) <br> | ||
| + | - 3g, -h were picked from plate L2.2a (30.07.10) <br><br> | ||
| + | ̈́* On these plates were many red colonies, this might be because no extraction was done after the cutting, which would result in un-cut plasmids to be pressent. <br> | ||
| + | |||
| + | '''''Pre-mix x 25''''' | ||
| + | <br> | ||
| + | 87.5 ul water <br> | ||
| + | 62.5 ul 10 x TAQ buffer <br> | ||
| + | 25 ul MgCl2 <br> | ||
| + | 25 ul VF2 <br> | ||
| + | 25 ul VR <br> | ||
| + | 12.5 ul 10mM dNTP <br> | ||
| + | 3 ul TAQ (we've got a new TAQ stock which is twice as strong as the previous, so now we only use 1/8 TAQ of the amount in the protocol. ((1/8 x 1 ul) x 24)) <br> | ||
| + | |||
| + | '''''PCR Program:''''' | ||
| + | <br> | ||
| + | <html> | ||
| + | <head> | ||
| + | <meta content="text/html; charset=ISO-8859-1" | ||
| + | http-equiv="content-type"> | ||
| + | <title></title> | ||
| + | </head> | ||
| + | <body> | ||
| + | <table style="text-align: left; width: 100px;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Progress<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">Temp (celcius)<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">Time (min) FlhDC<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">Time (min) DT<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Start<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">94<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">2<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">2<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Denaturing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">94<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">1<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">1<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Annealing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">55<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">1<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">1<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Elongation<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">72<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">1.5<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">0.5<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">GOTO 2<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;"><br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;"><br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;"><br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">End<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">72<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">3<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">3<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Hold<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">4<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;"><br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;"><br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <br> | ||
| + | </body> | ||
| + | </html> | ||
| + | <br> | ||
| + | <br> | ||
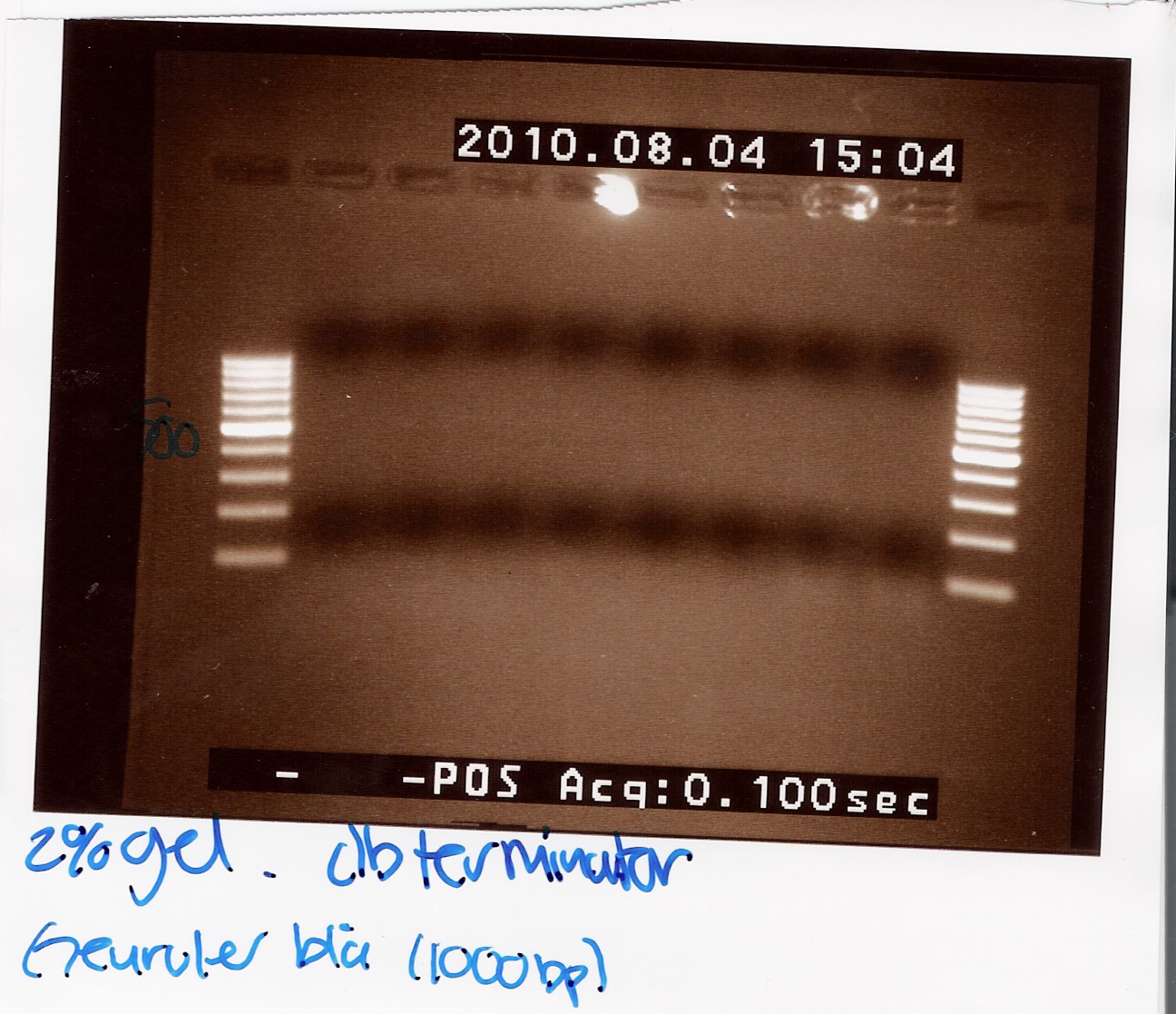
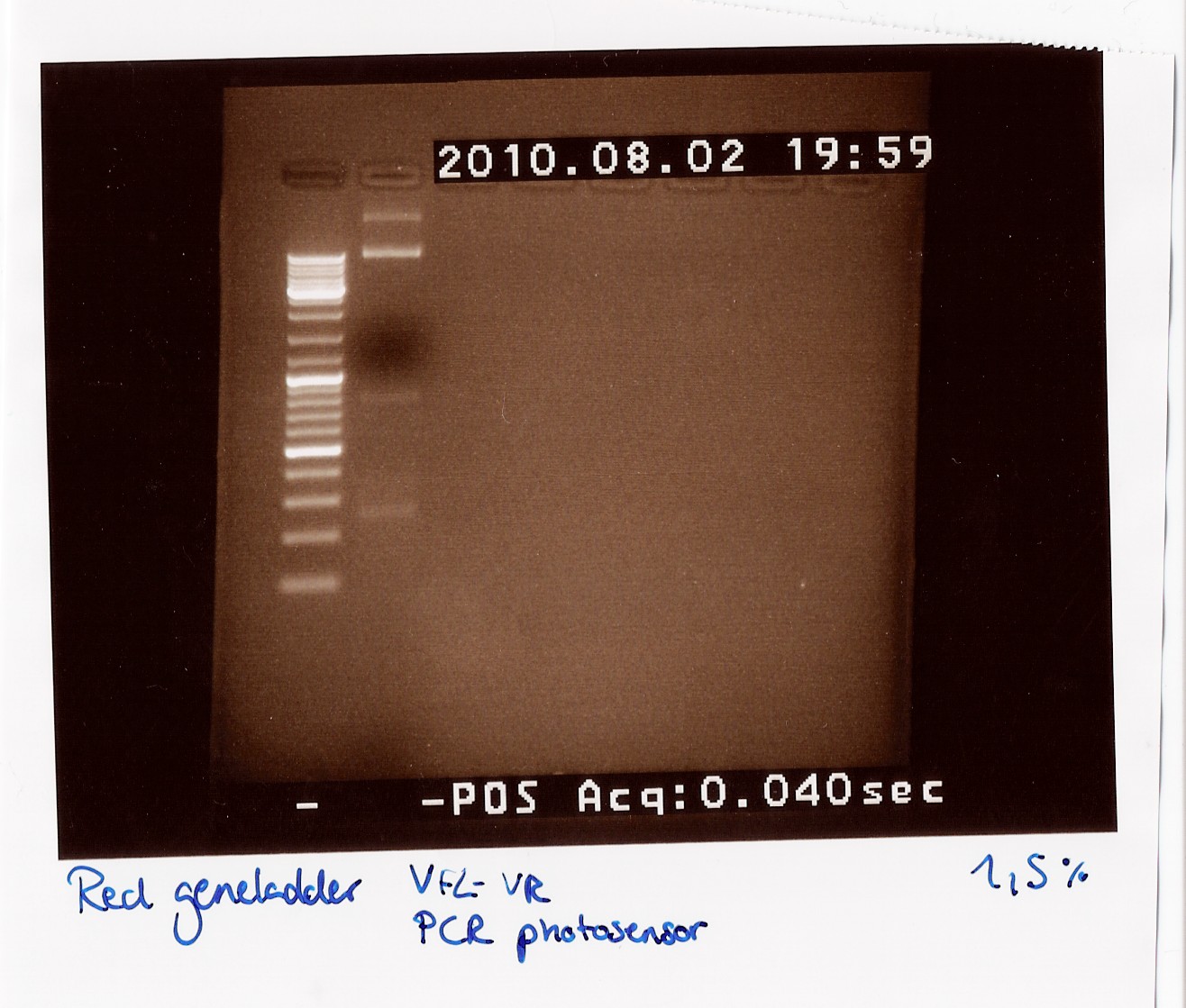
| + | ''Results:'' | ||
| + | <br> | ||
| + | '''''DT:''''' | ||
| + | <br> | ||
| + | The double terminator was run on a 2% gel with a 100-1000 ladder. The gel shows no DNA at all! | ||
| + | <br> | ||
| + | [[Image:Team-SDU Denmark Double terminator col. PCR.jpg|300px]] | ||
| + | <br><br> | ||
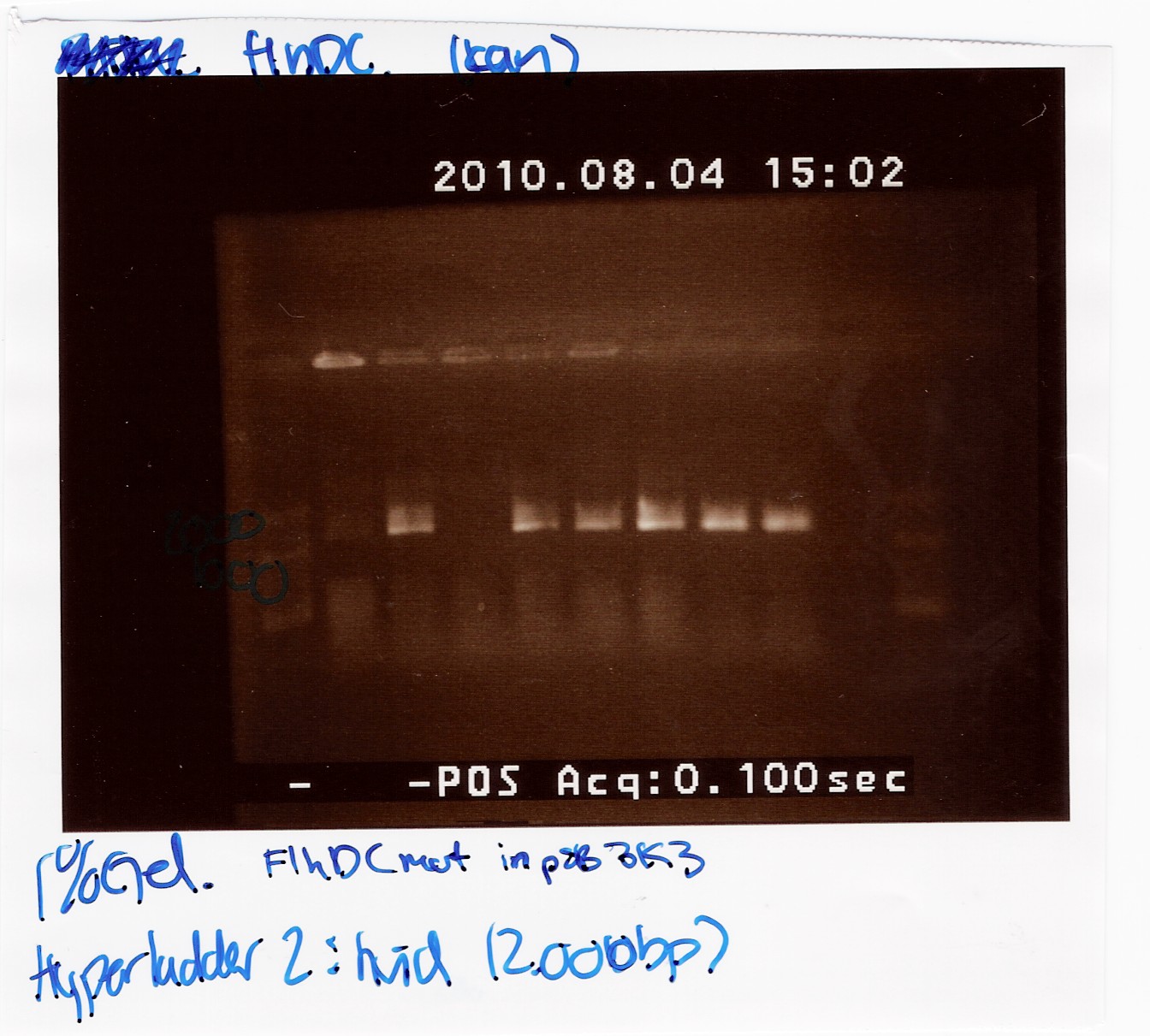
| + | '''''FlhDCmut in pSB3k3:'''''' | ||
| + | <br> | ||
| + | This was run on a 1% gel with a 50-2000bp ladder. The light in this picture is bad, but it does show high concentration of DNA in six of the eight ligations. The bands are positioned above 1000bp which is expected since the FlhDCmut gene with VF2-VR is 1248bp. <br> | ||
| + | [[Image:Team-SDU denmark FlhDCmut in psb3k3.jpg|300px]] | ||
| + | <br><br> | ||
| + | '''''FlhDCmut in pSB1c3:''''' | ||
== Group: Photosensor == | == Group: Photosensor == | ||
Revision as of 12:21, 5 August 2010
 "
"