Team:SDU-Denmark/labnotes4
From 2010.igem.org
(Difference between revisions)
(→Lab notes (7/19 - 7/25)) |
(→Lab notes (8/2 - 8/8)) |
||
| Line 7: | Line 7: | ||
__TOC__ | __TOC__ | ||
| + | |||
| + | == Group: Photosensor == | ||
| + | |||
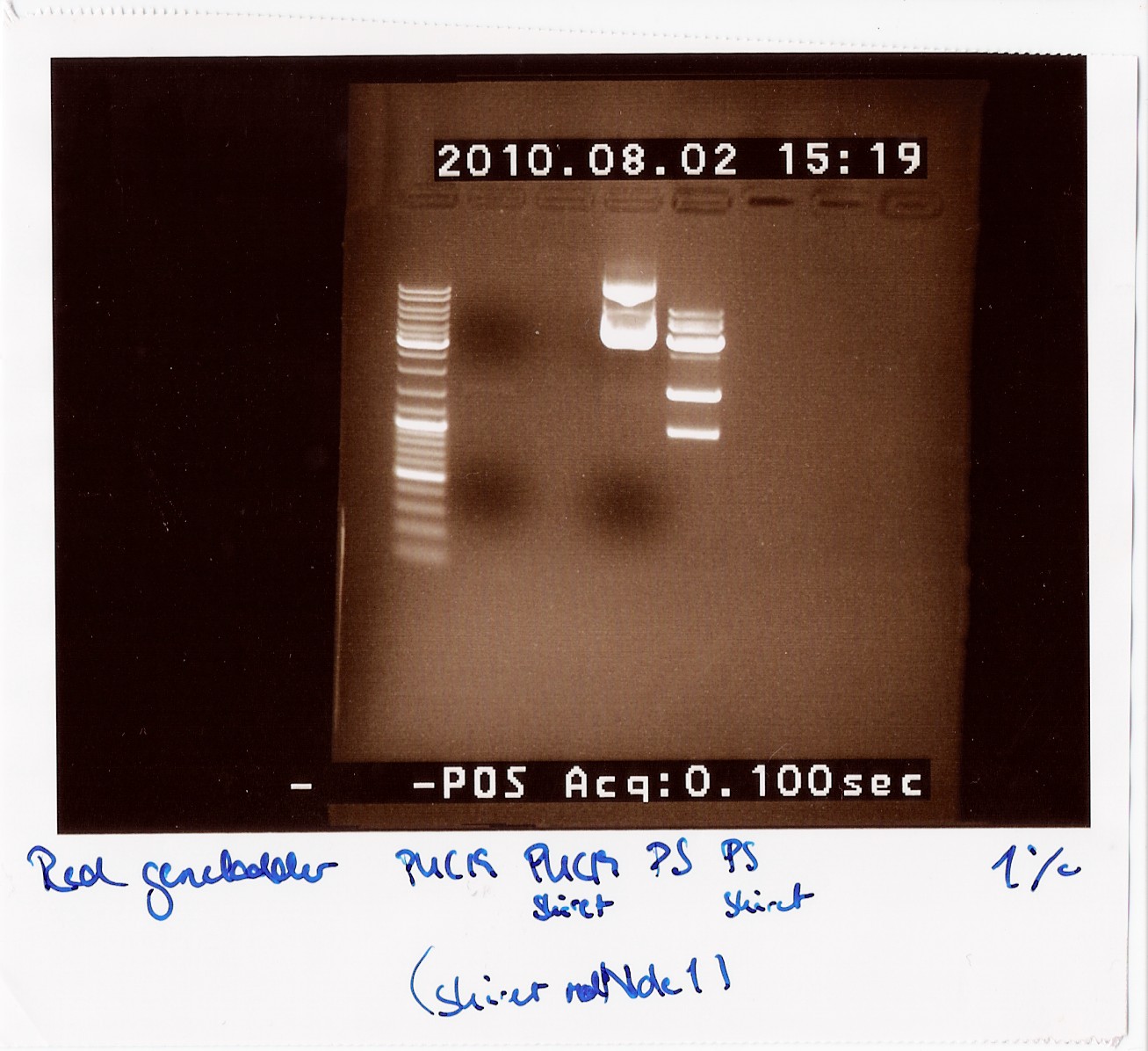
| + | === Restriction digest of PUC19 and the photosensor in pKJ606 with Nde1 === | ||
| + | Start date: 2/08 End date: 2/08<br> | ||
| + | ''Methods:'' Restriction digest, gel electrophoresis <br><br> | ||
| + | ''Protocol:''[https://2010.igem.org/Team:SDU-Denmark/protocols#RD1.1 RD1.1] <br><br> | ||
| + | ''Experiment done by:'' LC<br><br> | ||
| + | ''Notes:'' Since all PCR attempts with VF2 and VR had failed until now, we wanted to see if there was an insert at all in the physical DNA we received. Therefore we cut PUC19 and the photosensor to compare their lengths to each other, since the photosensor should be longer than PUC19 uncut and the cut photosensor should consist of two pieces, a 900 - 1000 BP piece and a 3600 BP piece. <br> | ||
| + | Loading order:<br> | ||
| + | 1: PUC19 uncut<br> | ||
| + | 2: PUC19 cut<br> | ||
| + | 3: Photosensor uncut<br> | ||
| + | 4: Photosensor cut<br><br> | ||
| + | ''Results:''<br> | ||
| + | [[Image:Team-SDU-Denmark-rd38.jpg|600px]]<br> | ||
| + | |||
| + | ''Analysis:''<br> | ||
| + | We can clearly see that the photosensor uncut is longer than the uncut PUC19. We can also see multiple bands on the cut photosensor, which are the expected lengths. This indicates that the photosensor gene should be inserted between VF2 and VR, so we are still unsure why the PCR is not working.<br> | ||
| + | |||
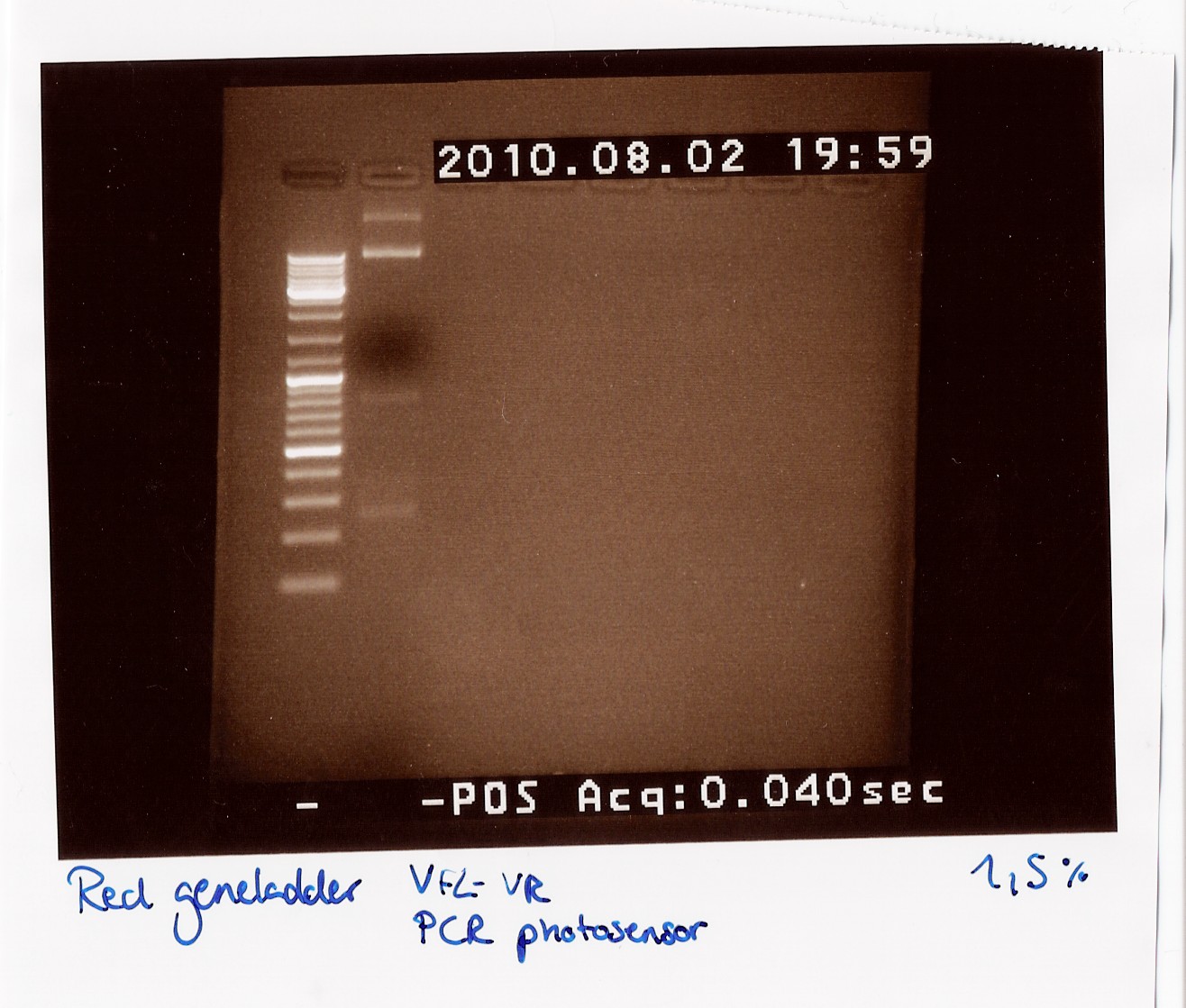
| + | === Dreamtaq Green PCR Master Mix (2X) PCR on photosensor with VF2 and VR === | ||
| + | Start date: 2/08 End date: 2/08<br> | ||
| + | ''Methods:'' PCR, gel electrophoresis <br><br> | ||
| + | ''Protocol:'' Dream TAQ Green PCR Master Mix protocol (modified) <br><br> | ||
| + | ''Experiment done by:'' LC<br><br> | ||
| + | ''Notes:'' Instead of a 50 µl reaction we ran a 25 µl reaction. It was mixed like this: <br> | ||
| + | <table style="text-align: left; height: 184px; width: 239px;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 169px;">Dream TAQ Master | ||
| + | Mix<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">12,5 µl<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 169px;">VF2<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">1 µl<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 169px;">VR<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">1 µl<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 169px;">Template<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">0,5 µl<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top; width: 169px;">Water<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 52px;">10 µl<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table><br> | ||
| + | We used a miniprep of the photosensor as template DNA, hence the small amount of template. The PCR program used was as follows:<br> | ||
| + | <table style="text-align: left; width: 338px; height: 272px;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tbody> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Step<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">Temperature<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 65px;">Time<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 38px;">Number of cycles<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Initial denaturation<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">95°<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 65px;">1 - 3 min<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 38px;">1<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Denaturation<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">95°<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 65px;">1 min<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 38px;">30<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Annealing<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">55°<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 65px;">1 min<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 38px;">30<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Extension<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">72°<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 65px;">3 min<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 38px;">30<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td style="vertical-align: top;">Final extension<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top;">72°<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 65px;">3 min<br> | ||
| + | </td> | ||
| + | <td style="vertical-align: top; width: 38px;">1<br> | ||
| + | </td> | ||
| + | </tr> | ||
| + | </tbody> | ||
| + | </table> | ||
| + | <br> | ||
| + | <br> | ||
| + | ''Results:''<br> | ||
| + | [[Image:Team-SDU-Denmark-dtaq28.jpg|600px]]<br> | ||
| + | |||
| + | ''Analysis:''<br> | ||
| + | We got bands at 270 BP, 850 BP (the distance between VF2 and VR without insert) and two bands very high up in the gel (probably the template which did not get amplified and could not run through the 1,5% gel).<br><br> | ||
Revision as of 07:46, 3 August 2010
 "
"