Team:SDU-Denmark/labnotes4
From 2010.igem.org
(Difference between revisions)
(→Insertion of B0015 in pSB3C5) |
|||
| (8 intermediate revisions not shown) | |||
| Line 33: | Line 33: | ||
<br> | <br> | ||
--[[User:Louch07|Louch07]] 10:20, 5 August 2010 (UTC) | --[[User:Louch07|Louch07]] 10:20, 5 August 2010 (UTC) | ||
| + | |||
| + | ==== Vol. 4 - Fourth time is the charme ==== | ||
| + | ''Done by:'' Pernille & Louise <br> | ||
| + | ''Date:'' August 5th <br> | ||
| + | ''Protocol:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#RD1.1 RD1.1] <br> | ||
| + | ''Notes:'' <br> | ||
| + | |||
| + | '''Cutting FlhDC''' <br> | ||
| + | tube 1: FlhDC uncut (freeze tube 5) <br> | ||
| + | tube 2: FlhDC cut with pst1 (5)<br> | ||
| + | tube 3: FlhDC uncut (freeze tube 21) <br> | ||
| + | tube 4: FlhDC cut with pst1 (21) <br> | ||
| + | <br> | ||
| + | '''Cutting of FlhDCmut in pSB3K3''' <br> | ||
| + | tube 5: FlhDCmut uncut (PCR product F, from august 4th) <br> | ||
| + | tube 6: FlhDCmut cut with pst1 (F) <br> | ||
| + | tube 7: FlhDCmut cut with spe1 (F) <br> | ||
| + | <br> | ||
| + | tube 8: FlhDCmut uncut (PCR product G, from august 4th) <br> | ||
| + | tube 9: FlhDCmut cut with pst1 (G) <br> | ||
| + | tube 10: FlhDCmut cut with spe1 (G) <br> | ||
| + | <br> | ||
| + | '''Cutting of FlhDCmut in pSB1C3''' <br> | ||
| + | tube 11: FlhDCmut uncut (PCR product C, from august 4th) <br> | ||
| + | tube 12: FlhDCmut cut with pst1 (C) <br> | ||
| + | tube 13: FlhDCmut cut with spe1 (C) <br> | ||
| + | <br> | ||
| + | tube 14: FlhDCmut uncut (PCR product D, from august 4th) <br> | ||
| + | tube 15: FlhDCmut cut with pst1 (D) <br> | ||
| + | tube 16: FlhDCmut cut with spe1 (D) <br> | ||
| + | <br><br> | ||
| + | '''Restriction mixture''' <br> | ||
| + | 13ul water <br> | ||
| + | 2ul FD green buffer <br> | ||
| + | 5ul PCR product <br> | ||
| + | 1ul enzyme <br><br> | ||
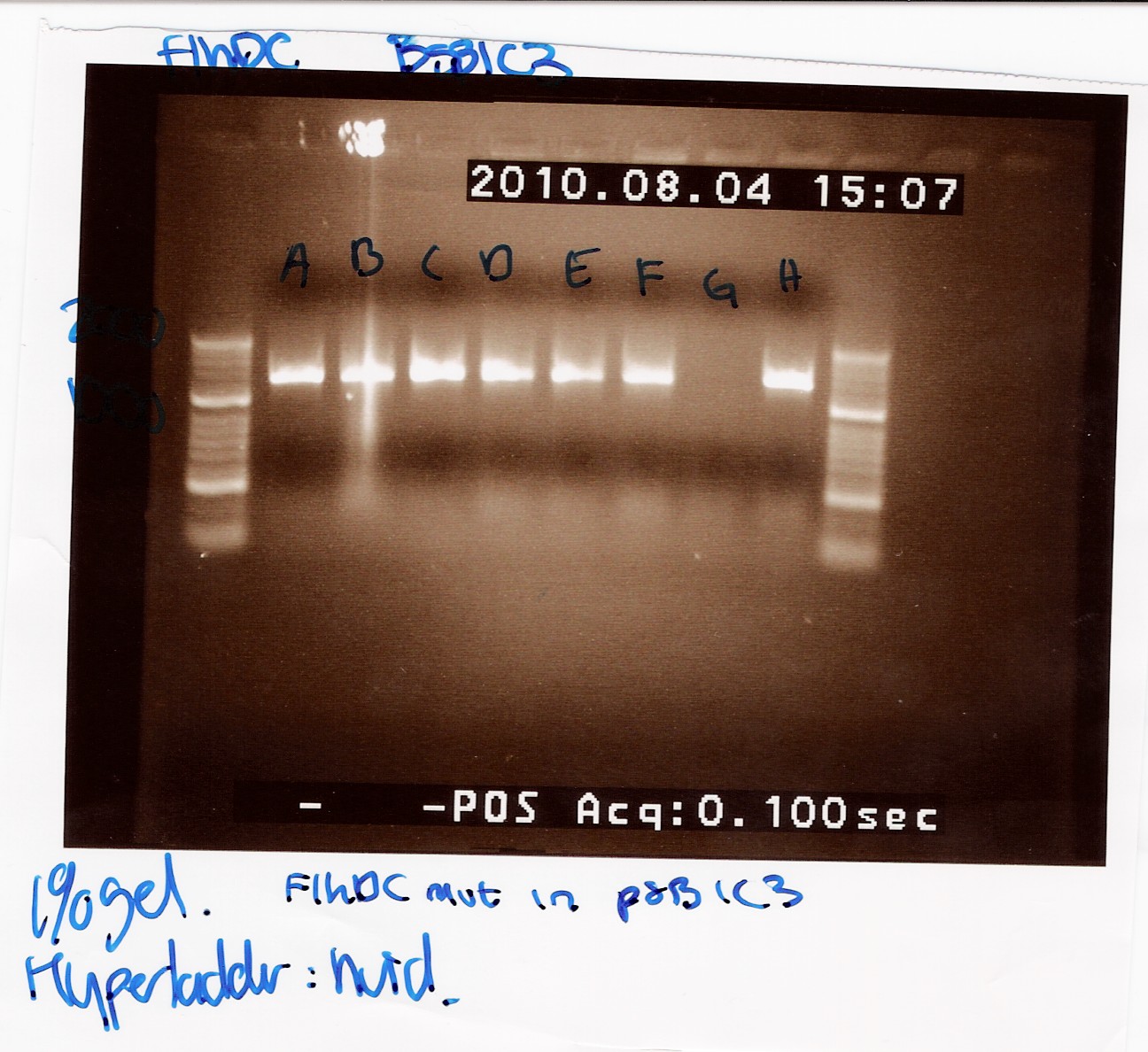
| + | ''Results:'' | ||
| + | <br> | ||
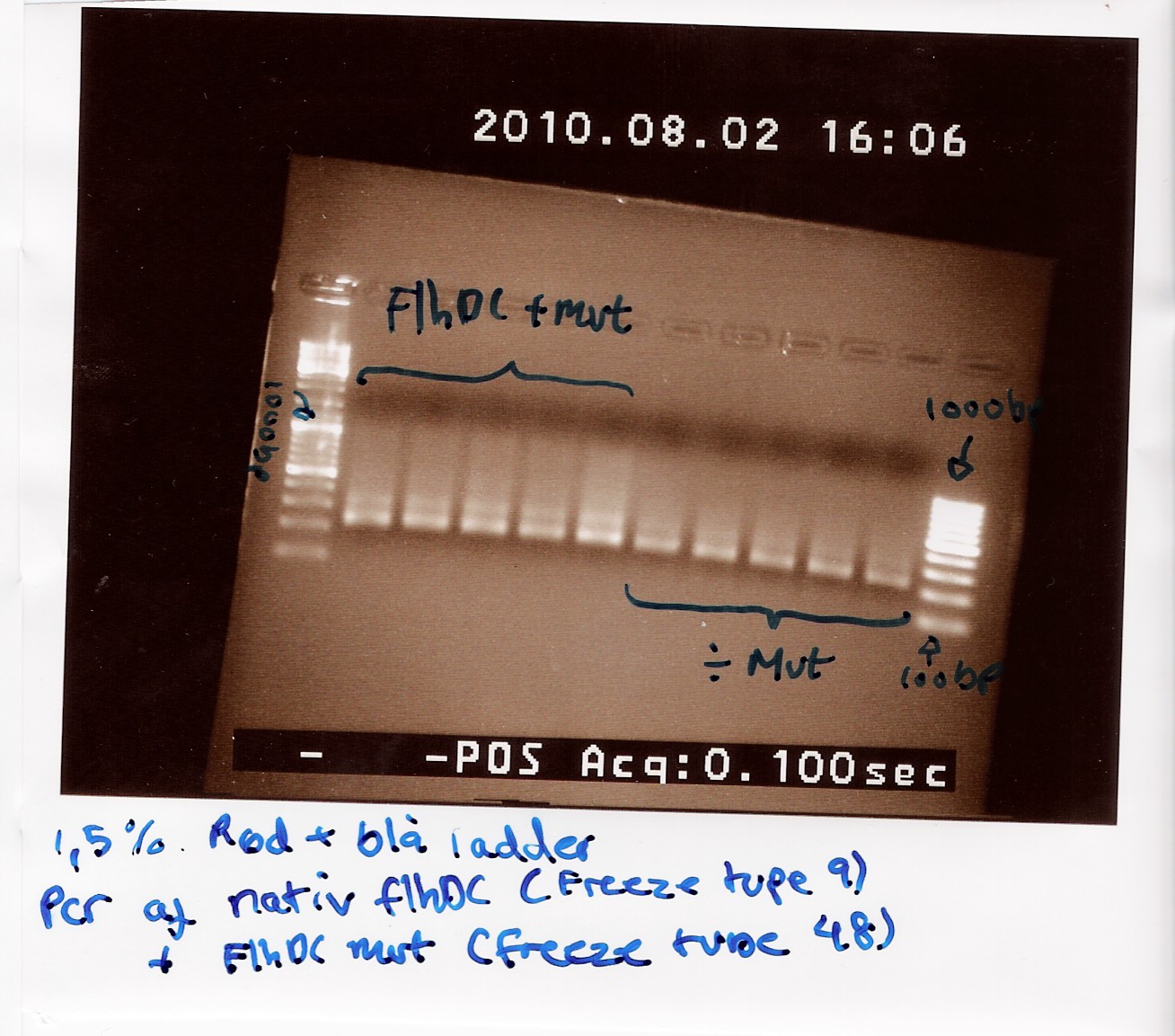
| + | In the picture U = Uncut, P = Pst1 and S = Spe1. The Native FlhDC has an internal Pst1 site, while this site has been altered in the FlhDCmut gene. As expected all the cut samples show two bands, while the uncut samples only show one band. The picture shows a difference between the cut band and the greatest cut band in the native FlhDC, the cut band is slightly leighter than the uncut (ca. 100bp). The FlhDCmut samples show no specefic difference between the uncut band and the greatest cut band. <br> | ||
| + | This indicates that a grater part of the native FlhDC is cut of than is cut of the FlhDCmut. <br> | ||
| + | [[Image:Team- SDU Denmark-FlhDCmut i 1C3 og 3K3 + nativ flhDCmut.jpg|300px]] | ||
=== Colony PCR of FlhDCmut and DT (B0015) === | === Colony PCR of FlhDCmut and DT (B0015) === | ||
| Line 177: | Line 218: | ||
<br><br> | <br><br> | ||
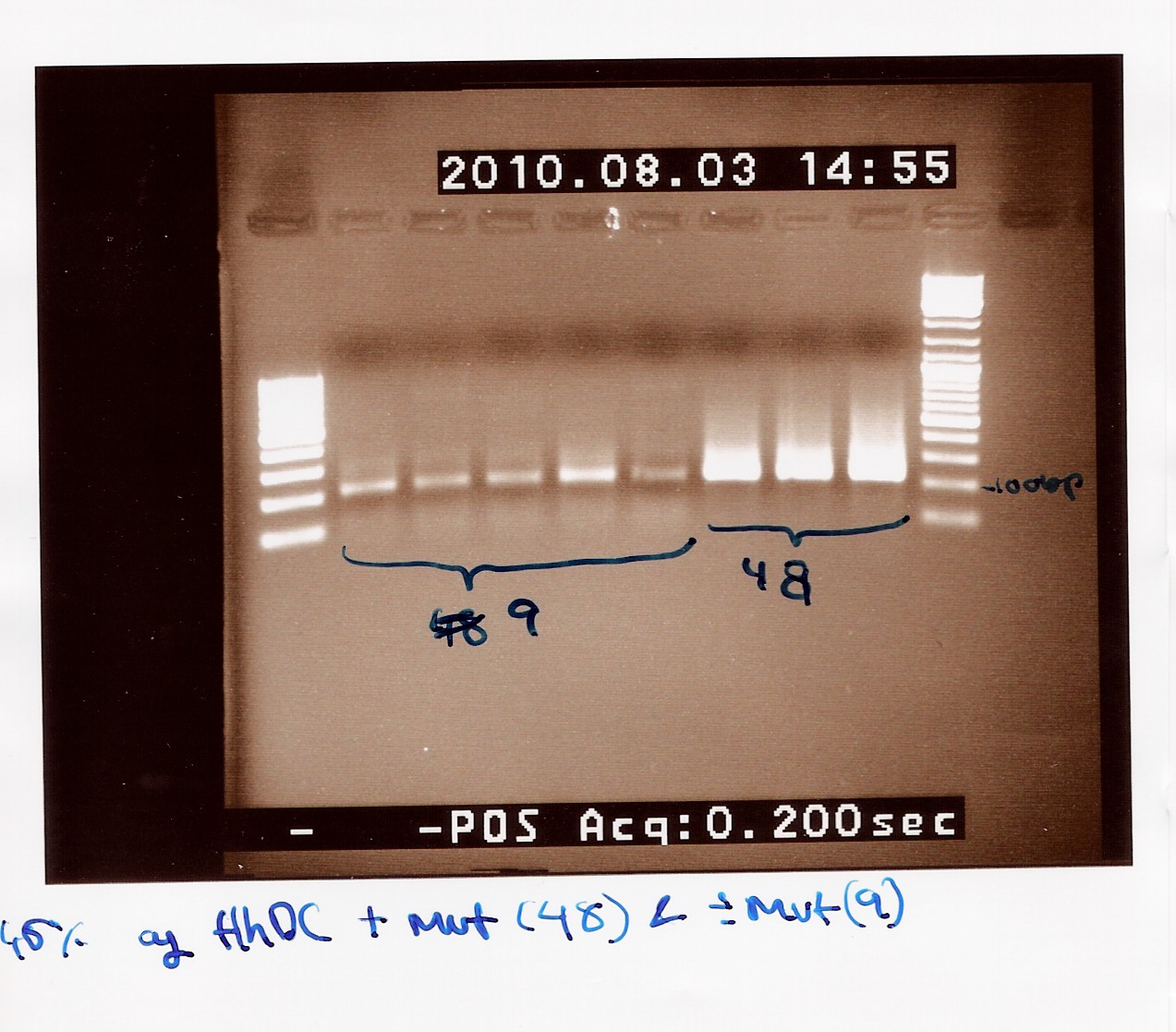
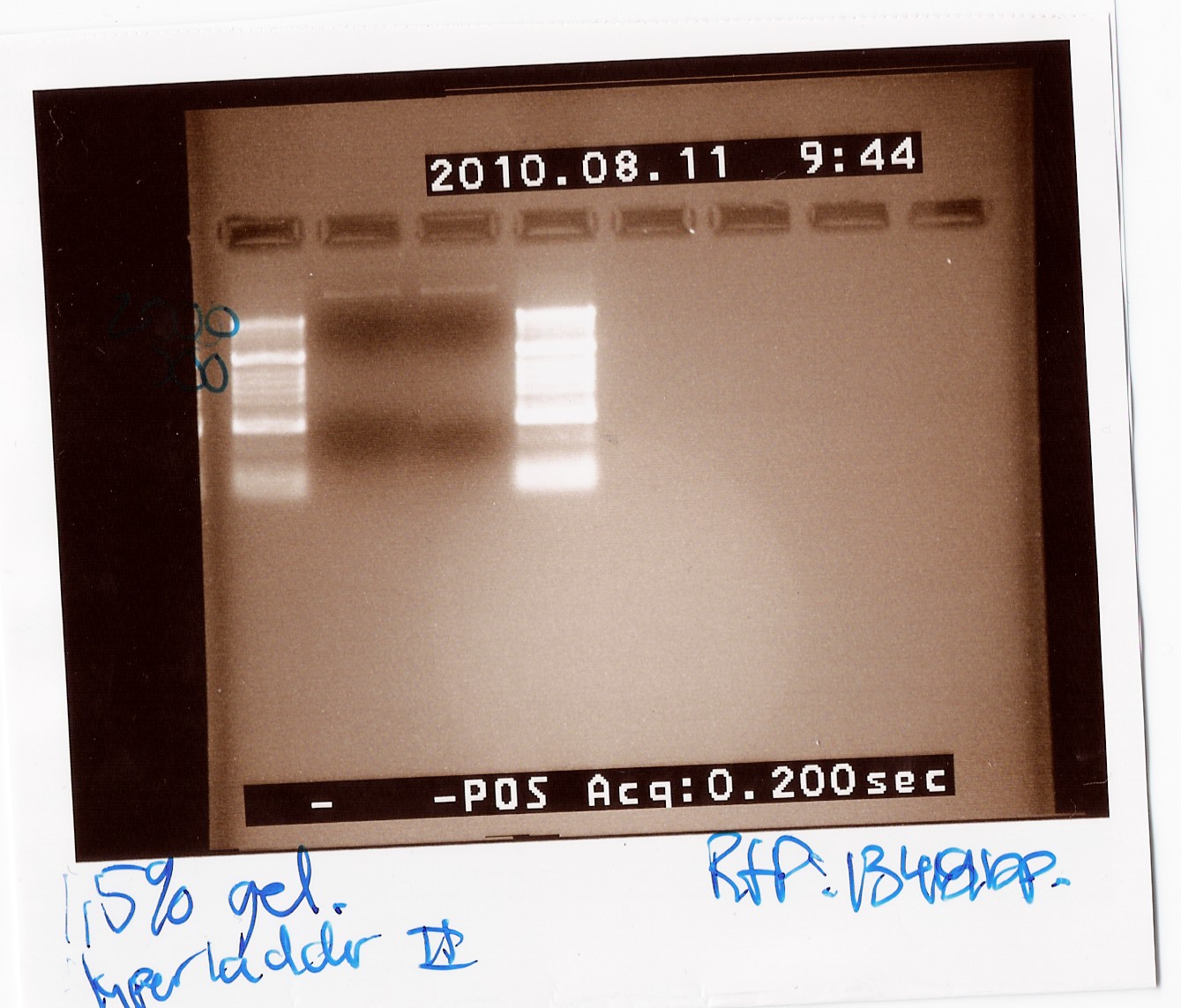
'''''FlhDCmut in pSB1c3:''''' | '''''FlhDCmut in pSB1c3:''''' | ||
| + | <br> | ||
| + | This was run on a 1% gel with a 50-2000bp ladder. The picture shows high concentration of DNA in seven of the eight ligations. The bands are positioned above 1000bp which is expected since the FlhDCmut gene with VF2-VR is 1248bp. <br> | ||
| + | [[Image:Team-SDU Denmark- FlhDCmut i pSB1C3 forkert mut.jpg|300px]] | ||
=== Colony PCR of pSB3T3 w RFP(J04450) === | === Colony PCR of pSB3T3 w RFP(J04450) === | ||
| Line 183: | Line 227: | ||
''Protocol:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1 CP1.1] <br> | ''Protocol:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.1 CP1.1] <br> | ||
''Notes:'' The experiment was done with Taq will the purpose only was to check if the RFP was amplificated when using the primers VR og VF2. The PRC program used was c.f. the protocol but the elongation time was 1min and 30 sec while the biobrick in the pSB3T3 plamid are 1319bp long. For the PCR i used the mini prep product from freeze tube 29. <br> | ''Notes:'' The experiment was done with Taq will the purpose only was to check if the RFP was amplificated when using the primers VR og VF2. The PRC program used was c.f. the protocol but the elongation time was 1min and 30 sec while the biobrick in the pSB3T3 plamid are 1319bp long. For the PCR i used the mini prep product from freeze tube 29. <br> | ||
| - | ''Results:''<br> [[Image:Team-SDU-Denmark-psb3T3wRFP.jpg | | + | ''Results:''<br> [[Image:Team-SDU-Denmark-psb3T3wRFP.jpg | 300px]]<br> |
Unfortunately there was no band around 1400bp and therefore the amplification was not succeed. the upper band is probably the plamid. | Unfortunately there was no band around 1400bp and therefore the amplification was not succeed. the upper band is probably the plamid. | ||
--[[User:Pernm07|Pernm07]] 08:58, 11 August 2010 (UTC) | --[[User:Pernm07|Pernm07]] 08:58, 11 August 2010 (UTC) | ||
| Line 204: | Line 248: | ||
2 uL PCR product of B0015 (no. 43 white) was used as template for each PCR reaction.3 PCR reactions were prepared.<br> | 2 uL PCR product of B0015 (no. 43 white) was used as template for each PCR reaction.3 PCR reactions were prepared.<br> | ||
Premix for 4 PCR reactions:<br> | Premix for 4 PCR reactions:<br> | ||
| - | <table style="text-align: left; width: | + | <table style="text-align: left; width: 300px;" border="1" |
cellpadding="2" cellspacing="2"> | cellpadding="2" cellspacing="2"> | ||
<tr> | <tr> | ||
| Line 234: | Line 278: | ||
48uL premix is distrubuted into each PCR tube. PCR tubes are marked B0015.A-C<br> | 48uL premix is distrubuted into each PCR tube. PCR tubes are marked B0015.A-C<br> | ||
PCR program:<br> | PCR program:<br> | ||
| - | <table style="text-align: left; width: | + | <table style="text-align: left; width: 300px;" border="1" |
cellpadding="2" cellspacing="2"> | cellpadding="2" cellspacing="2"> | ||
<tr> | <tr> | ||
| Line 284: | Line 328: | ||
purified B0015 PCR product and mini prep of pSB3C5 (28 white) are digested with EcoRI and PstI.<br> | purified B0015 PCR product and mini prep of pSB3C5 (28 white) are digested with EcoRI and PstI.<br> | ||
Restriction mixture:<br> | Restriction mixture:<br> | ||
| - | <table style="text-align: left; width: | + | <table style="text-align: left; width: 300px;" border="1" |
cellpadding="2" cellspacing="2"> | cellpadding="2" cellspacing="2"> | ||
<tr> | <tr> | ||
| Line 331: | Line 375: | ||
45ng of vector was used for each ligation.<br> | 45ng of vector was used for each ligation.<br> | ||
Ligation mixtures:<br> | Ligation mixtures:<br> | ||
| - | <html><table style="text-align: left; width: | + | <html><table style="text-align: left; width: 300px;" border="1" |
cellpadding="2" cellspacing="2"> | cellpadding="2" cellspacing="2"> | ||
<tr> | <tr> | ||
| Line 378: | Line 422: | ||
For compotent cells a Top 10 E.coli coloni was inoculated in 5mL LB media ON. <br> | For compotent cells a Top 10 E.coli coloni was inoculated in 5mL LB media ON. <br> | ||
ON culture was diluted and grown to reach exponential phase.<br> | ON culture was diluted and grown to reach exponential phase.<br> | ||
| - | <table style="text-align: left; width: | + | <table style="text-align: left; width: 200px;" border="1" |
cellpadding="2" cellspacing="2"> | cellpadding="2" cellspacing="2"> | ||
<tr> | <tr> | ||
| Line 408: | Line 452: | ||
''Analysis:''<br> | ''Analysis:''<br> | ||
The transformation appears to have been successful and white colonies where selected for coloni PCR.<br><br> | The transformation appears to have been successful and white colonies where selected for coloni PCR.<br><br> | ||
| + | |||
| + | ==== Coloni PCR ==== | ||
| + | ''Date:'' 5/8<br> | ||
| + | ''Protocols:'' [https://2010.igem.org/Team:SDU-Denmark/protocols#CP1.3 CP1.3]<br><br> | ||
| + | ''Notes:''<br> | ||
| + | 8 white colonies were selected from L1, L2, and L3 plates repectively.<br> | ||
| + | Premix x 10:<br> | ||
| + | <table style="text-align: left; width: 300px;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td>25uL</td> | ||
| + | <td>taq buffer (+KCl - MgCl2)</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10uL</td> | ||
| + | <td>MgCl2</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10uL</td> | ||
| + | <td>VF2</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>10uL</td> | ||
| + | <td>VR</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>5uL</td> | ||
| + | <td>d'NTP</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>35uL</td> | ||
| + | <td>H2O</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>1uL</td> | ||
| + | <td>taq polymerase enzyme</td> | ||
| + | </tr> | ||
| + | </table><br> | ||
| + | PCR tubes are marked Lig3.A-H<br> | ||
| + | PCR program:<br> | ||
| + | <table style="text-align: left; width: 300px;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td>Start</td> | ||
| + | <td>94C</td> | ||
| + | <td>2min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Denaturating</td> | ||
| + | <td>94C</td> | ||
| + | <td>1min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Annealing</td> | ||
| + | <td>55C</td> | ||
| + | <td>1min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Elongation</td> | ||
| + | <td>72C</td> | ||
| + | <td>30s</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Go to</td> | ||
| + | <td>2</td> | ||
| + | <td>rep. 29x</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>End</td> | ||
| + | <td>72C</td> | ||
| + | <td>3min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Hold</td> | ||
| + | <td>4C</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table><br> | ||
| + | PCR product was loaded onto a 2% agarose gel. Gene ruler 100bp DNA ladder (blue) was used as marker.<br><br> | ||
| + | ''Results:'' | ||
| + | <br><br> | ||
| + | ''Analysis:''<br> | ||
| + | No visible results. Do-over!!<br><br> | ||
== Group: Photosensor == | == Group: Photosensor == | ||
| Line 423: | Line 550: | ||
4: Photosensor cut<br><br> | 4: Photosensor cut<br><br> | ||
''Results:''<br> | ''Results:''<br> | ||
| - | [[Image:Team-SDU-Denmark-rd38.jpg| | + | [[Image:Team-SDU-Denmark-rd38.jpg|300px]]<br> |
''Analysis:''<br> | ''Analysis:''<br> | ||
| Line 535: | Line 662: | ||
<br> | <br> | ||
''Results:''<br> | ''Results:''<br> | ||
| - | [[Image:Team-SDU-Denmark-dtaq28.jpg| | + | [[Image:Team-SDU-Denmark-dtaq28.jpg|300px]]<br> |
''Analysis:''<br> | ''Analysis:''<br> | ||
We got bands at 270 BP, 850 BP (the distance between VF2 and VR without insert) and two bands very high up in the gel (probably the template which did not get amplified and could not run through the 1,5% gel).<br><br> | We got bands at 270 BP, 850 BP (the distance between VF2 and VR without insert) and two bands very high up in the gel (probably the template which did not get amplified and could not run through the 1,5% gel).<br><br> | ||
| + | |||
| + | === pfu PCR of photosensor === | ||
| + | ''Date:'' 10/8<br> | ||
| + | ''Experiment done by:'' Maria<br> | ||
| + | ''Methods:'' PCR, gel electrophoresis<br> | ||
| + | ''Notes:'' <br> | ||
| + | Miniprep of photosensor in pJK606 was used as template. 2uL template was used for each reaction.<br> | ||
| + | 4 PCR reaction was prepared.<br> | ||
| + | Premix x 5: <br> | ||
| + | <table style="text-align: left; width: 300px;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td>25uL</td> | ||
| + | <td>10 x pfu</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>7.5uL</td> | ||
| + | <td>VF2</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>7.5uL</td> | ||
| + | <td>VR</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>7.5uL</td> | ||
| + | <td>d'NTP</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>210uL</td> | ||
| + | <td>H2O</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>2uL</td> | ||
| + | <td>pfu polymerase enzyme</td> | ||
| + | </tr> | ||
| + | </table><br> | ||
| + | 48uL premix is distributed into each PCR tube. PCR tubes are marked PS3.A-D.<br> | ||
| + | PCR program:<br> | ||
| + | <table style="text-align: left; width: 300px;" border="1" | ||
| + | cellpadding="2" cellspacing="2"> | ||
| + | <tr> | ||
| + | <td>Start</td> | ||
| + | <td>94C</td> | ||
| + | <td>3min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Denaturating</td> | ||
| + | <td>94C</td> | ||
| + | <td>2min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Annealing</td> | ||
| + | <td>55C</td> | ||
| + | <td>30s</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Elongation</td> | ||
| + | <td>72C</td> | ||
| + | <td>4min 30s</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Go to</td> | ||
| + | <td>2</td> | ||
| + | <td>rep. 29x</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>End</td> | ||
| + | <td>72C</td> | ||
| + | <td>2min</td> | ||
| + | </tr> | ||
| + | <tr> | ||
| + | <td>Hold</td> | ||
| + | <td>4C</td> | ||
| + | <td></td> | ||
| + | </tr> | ||
| + | </table><br> | ||
| + | PCR product are loaded onto a 1% gel and run at 130V. Gene ruler green was used as marker.<br><br> | ||
| + | ''Results:''<br> | ||
| + | <br> | ||
| + | ''Analysis:''<br> | ||
| + | No usable results.<br><br> | ||
| + | |||
| + | |||
</div> | </div> | ||
<div id="rightcolumn">What up!</div> | <div id="rightcolumn">What up!</div> | ||
Latest revision as of 21:43, 23 October 2010
 "
"