Team:SDU-Denmark/labnotes3
From 2010.igem.org
(→DNA extraction from gel of FlhDC mut) |
(→Follow-up colony PCR =) |
||
| Line 736: | Line 736: | ||
All samples were incubated at 17 degrees until they were used for transformation.<br><br> | All samples were incubated at 17 degrees until they were used for transformation.<br><br> | ||
| - | == Follow-up colony PCR === | + | === Follow-up colony PCR === |
Date: 26/7<br> | Date: 26/7<br> | ||
Methods: Colony PCR<br> | Methods: Colony PCR<br> | ||
| Line 1,134: | Line 1,134: | ||
''Protocol:'' [[https://2010.igem.org/Team:SDU-Denmark/protocols#RD1.1 RD1.1]] and gelpurificaton (DE1.3)<br><br> | ''Protocol:'' [[https://2010.igem.org/Team:SDU-Denmark/protocols#RD1.1 RD1.1]] and gelpurificaton (DE1.3)<br><br> | ||
''Notes:''because the concentration for both plasmid and biobrick is rather small I dobbelt the volumen of the reagent compared to the protocol. in the soultion contaning the brick the volumen of the PCR product was triple and to adjust the total volumen i added 5ul less water. We made 2 Restriction mixtures which both were 4 times the mixture in the protocol. both the plasmid and brick was cut at the E and P site. After the cutting the plasmid and brick was run on seperate gels because of there different size. the plasmid on a 1,5% gel and brick on a 2% agarose gel. When I observed the gel under the UV lamp the was no visible band on the gel were the brick was loaded. therefore it was only possible to do DNA extrated from the gel contraining the plasmid. The Dna was eluted in 10ul of water and I eluted two times. The DNA concentration was measured on the NaonDrop and was found to 55,41ng/ul for the first elution and 28,03ng/ul for the second. the to samples are pooled and stored in a freeze tube.<br><br> | ''Notes:''because the concentration for both plasmid and biobrick is rather small I dobbelt the volumen of the reagent compared to the protocol. in the soultion contaning the brick the volumen of the PCR product was triple and to adjust the total volumen i added 5ul less water. We made 2 Restriction mixtures which both were 4 times the mixture in the protocol. both the plasmid and brick was cut at the E and P site. After the cutting the plasmid and brick was run on seperate gels because of there different size. the plasmid on a 1,5% gel and brick on a 2% agarose gel. When I observed the gel under the UV lamp the was no visible band on the gel were the brick was loaded. therefore it was only possible to do DNA extrated from the gel contraining the plasmid. The Dna was eluted in 10ul of water and I eluted two times. The DNA concentration was measured on the NaonDrop and was found to 55,41ng/ul for the first elution and 28,03ng/ul for the second. the to samples are pooled and stored in a freeze tube.<br><br> | ||
| + | |||
== Group: Retinal == | == Group: Retinal == | ||
=== Colony PCR and extraction of ninaB gene from transformants === | === Colony PCR and extraction of ninaB gene from transformants === | ||
Revision as of 07:21, 16 August 2010
Lab notes (7/26 - 8/1)
Group: Flagella
Incertion of Promoter + RBS in pSB3T5
Done by: Christian and Louise
Parts used: J13002 (promoter+RBS) and pSB3T5
Restriction and Gel extraction
Protocol: [RD1.1]
Notes: We made 2 Restriction mixtures which both were 4 times the mixture in the protocol. We also added 10 ul less water and 10 ul more PCR product than discribed in the protocol
Restriction mixture
- 38 ul Water
- 4 ul EcoRI enzyme
- 4 ul PstI enzyme
- 8 ul Fast Digest green buffer
- 30 ul PCR product (Freeze tube white 25 PROMOTER + RBS)
OR
- 30 ul PCR product (Freeze tube white 29 PLASMID)
Loading:
2 x 21 ul J13002 was loaded on a 2% gel with a 100-1000bp ladder. The Restricted J13002 is 95bp.
2 x 21 ul pSB3T5 was loaded on a 1.5% gel with a 100-10,000 ladder. The restristricted plasmid is 3215bp.
Result:
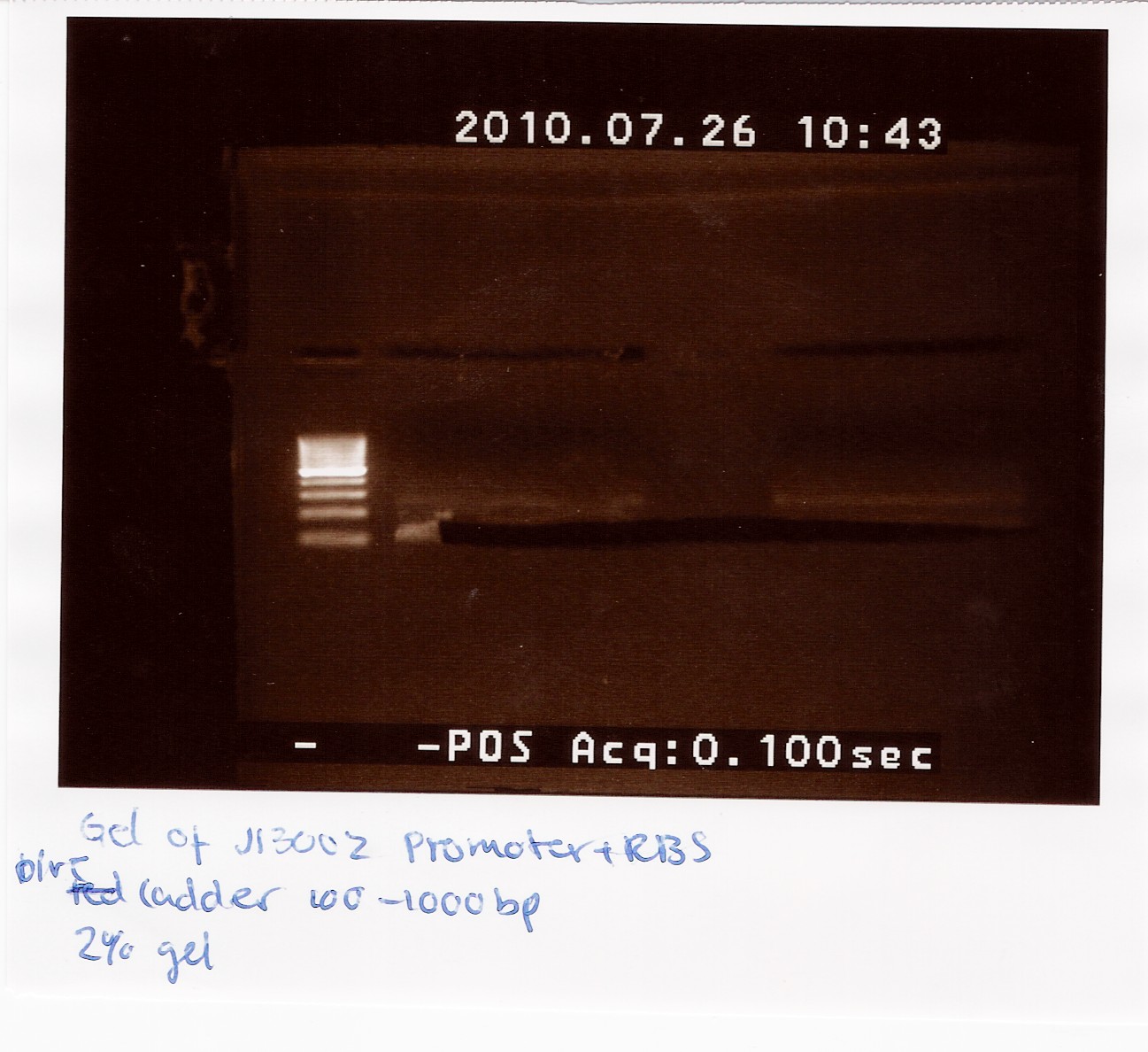
The picture shows a band around 100bp.
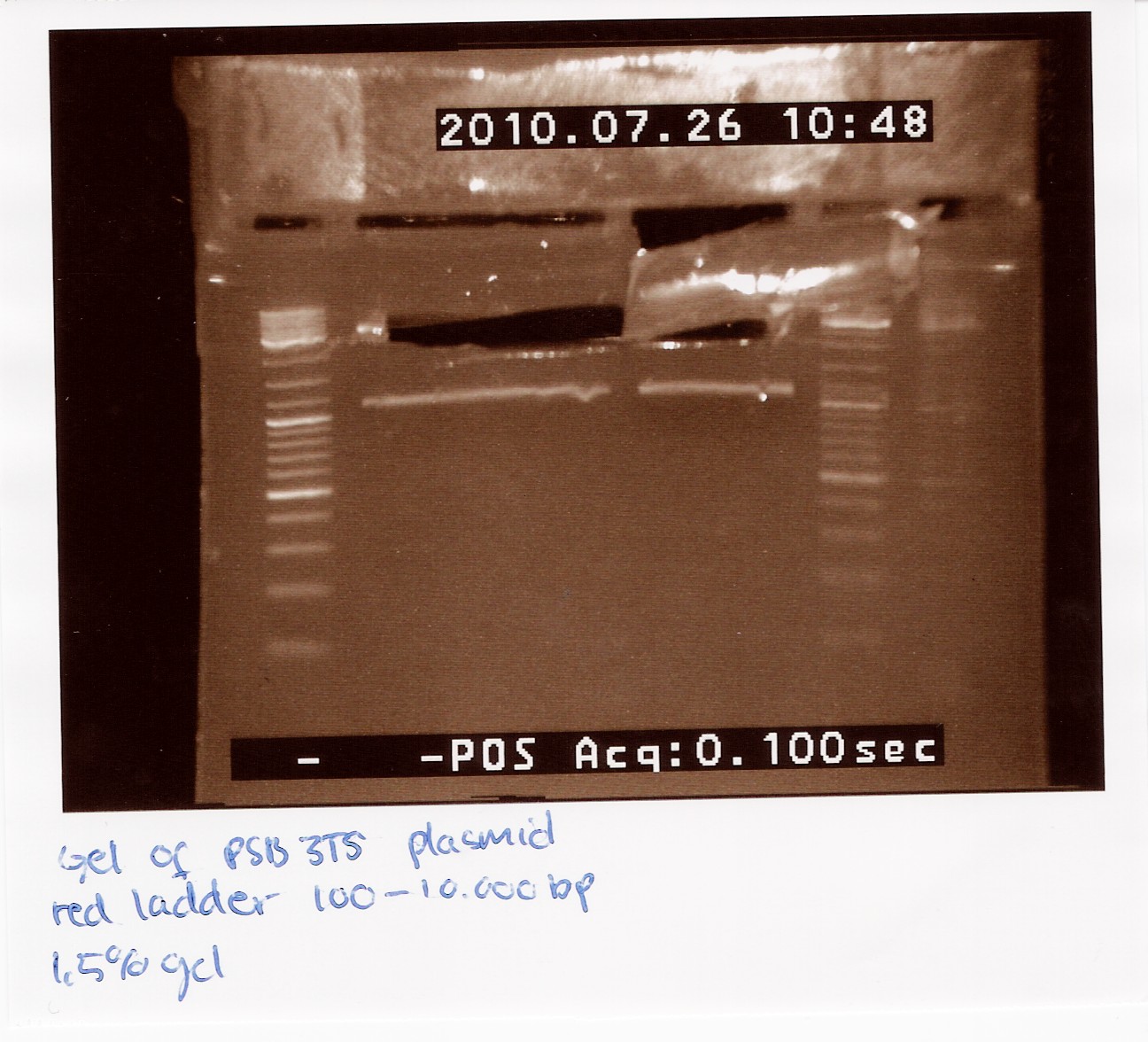
The picture showes two bands, one around 3000bp which is the plasmid and one around 1000bp, which is RFP.
Purification of J13002 and pSB3T5
Protocol:
Notes: four tubes were marked and weighed (all had a mass of 1g). Bands were cut from the restriction gels, added to the tubes and weighed.
| Tubes |
1P (Plasmid) |
2P |
3B (Brick) |
4B |
| Tube weight |
1g |
1g |
1g |
1g |
| Gel weight |
170mg |
110mg |
245mg |
160mg |
Sample capture: 300 ul Capture buffer type 3 was added to the tubes and the tubes were placed at 60 degrees for 20 minutes.
Sample binding:The Capture buffer sample mix were added to MicroSpin Columns, stood for 1 minute and centrifuged for 30 sec. at 16,000g.
The Wash and Dry was carried out according to the protocol.
Elution: 10 ul Elution buffer type 6 was added to the columns, stood for 1 minute and centrifuged for 1 minute at 16,000g. The columns were thrown out and NanoDroped.
NanoDrop:
| Tube |
Concentration (ng/ul) |
| 1P |
9.6 |
| 2P |
2.9 |
| 3B |
6.7 |
| 4B |
3.2 |
The Concentrations were quite low, but we pooled them (1P+2P and 3B+4B) and used them for ligation.
Pfu PCR of J13002 and B0015
Done by: Louise
Date: July 28th
Protocol: [CP1.1]
Notes:
5 x J13002 and 5 x B0015 were made.
Pre-mix x 10
400 ul water
50 ul Pfu-buffer + MgSO4
15 ul dNTP
15 ul VF2 primer
15 ul VR primer
5 ul Pfu polymerase
Total volume: 500 ul
To each PCR-tube 50 ul pre-mix and 2 ul DNA was mixed and the tubes were loaded on the PCR maschine.
PCR program
| Progress |
Temp. (c elcius) |
Time (min.) |
| Start |
94 |
3 |
| Denaturing |
94 |
2 |
| Annealing |
55 |
0.5 |
| Elongation |
72 |
0.5 |
| GOTO |
rep x 29 |
|
| End |
72 |
2 |
| Hold |
4 |
Result:
Nanodrop:
| Brick |
Conc. (ng/ul) |
260/280 |
260/230 |
| B0015 |
487.6 |
1.47 |
0.93 |
| J13002 |
490.7 |
1.44 |
0.94 |
The PCR results were run on a gel together with The FlhDC mut PCR result. The gel picture can be seen in the notes from FlhDC mut PCR.
The picture shows no expression of the Double terminator (B0015) and fine expression of Promoter + RBS (J13002). The Double terminator product was not used further, while the promnoter + RBS was run on a gel and extracted (described in notes below.
--Louch07 11:38, 28 July 2010 (UTC)
Pfu PCR of FlhDC with mutation
Done by: Louise
Date: July 28th
Protocol: [CHP1.1]
Notes: Template sample: Freeze tube white 31.
Six samples were made.
Pre-mix x 6
240 ul water
30 ul Pfu-buffer + MgSO4
9 ul 10mM dNTP mix
9 ul FlhD fw primer
9 ul FlhC rev primer
3 ul Pfu polymerase
Total volume: 300 ul
In the PCR tubes 50 ul pre-mix and 2 ul template was mixed and loaded into the PCR machine.
PCR Program:
| Progress |
Temp. (c elcius) |
Time (min.) |
| Start |
94 |
3 |
| Denaturing |
94 |
2 |
| Annealing |
55 |
0,5 |
| Elongation |
72 |
1 |
| GOTO |
rep x 29 |
|
| End |
72 |
2 |
| Hold |
4 |
Results:
NanoDrop:
| Brick |
Conc. (ng/ul) |
260/280 |
260/230 |
| FlhDC mut |
395.7 |
1.41 |
0.78 |
Gel electrophoresis:

The image shows a 2% gel where a 100-1000bp marker was loaded together with FlhDC mut, Double terminator and Promoter+RBS. 5 ul sample and 5 ul loading dye was loaded.
The FlhDC mut lane shows 4 bands. The heaviest band is about 1000bp which consists with the FlhDC gene length. The three other bands are between 100 and 250bp which means they are too small to be FlhD or FlhC fragments (ca. 400-500bp) and they are too big to be primers.
The FlhDC mut PCR samples were pooled and all of it was run on an extraction gel.
--Louch07 11:28, 28 July 2010 (UTC)
DNA extraction from gel of FlhDC mut
Done by: Louise
Date: July 29th
Protocol: [DE1.3]
Notes:
300 ul FlhDC mut PCR sample was loaded on a 2% gel with a 100-1000bp ladder.
6 microcentrifuge tubes were weighed without and with gel.
| Tube |
Weight (empty) |
Weight (w. gel) |
Weight (gel) |
| 1 |
0.97g |
1.18g |
0.2g |
| 2 |
0.99g |
1.13g |
0.14g |
| 3 |
0.98g |
1.21g |
0.23g |
| 4 |
0.98g |
1.05g |
0.07g |
| 5 |
0.98g |
1.14g |
0.16g |
| 6 |
0.97g |
1.08g |
0.11g |
Extraction gel:
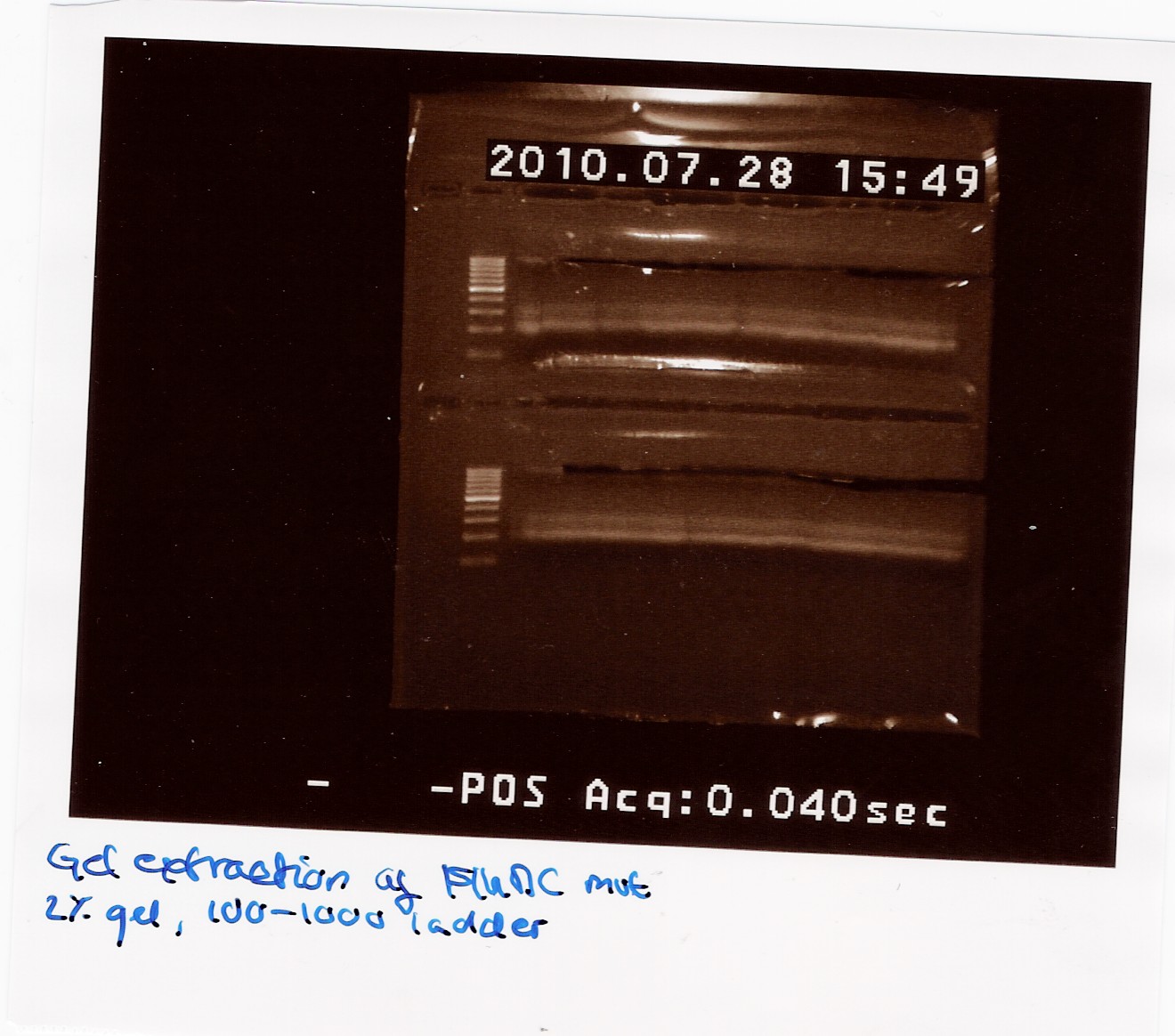
The gel was processed according to the protocol.
600 ul Capture buffer was used to dissolve the gel. This was transfered to 12 columns.
15 ul Elution buffer was used per. tube.
The final samples were NanoDropped (data not showen) and pooled. The pooled sample was NanoDropped: Conc: 2.38ng/ul. This is a low concentration.
This sample has freeze tube no. 44.
--Louch07 09:06, 29 July 2010 (UTC)
compent cell and transformation of photosensor and promotor+RNA in pSB3T5
Done by: Maria and Pernille
Date: July 27th
Protocol: CC1.1 and TR1.1
Notes:
Results: The OD of the cells were measured every hour and the corelation was:
| Time /h |
OD550 /Abs |
| 1 |
0,024 |
| 2 |
0,048 |
| 3 |
0,116 |
| 4 |
0,58 |
PCR of FlhDC freeze tube 23
Done by: Louise
Date: july 29th
Protcol: CP1.1
Notes: Since the PCR of freeze tube 31 shoved pour results on the gel, we are now trying another sample (freeze tube 23) This is made from sample 14 and 16 (mutation PCR samples). The gel picture from these samples showed the strongest band in the samples run with annealing temperature of 64.5 celcius.
Results: The PCR result was pooled in freeze tube 49.
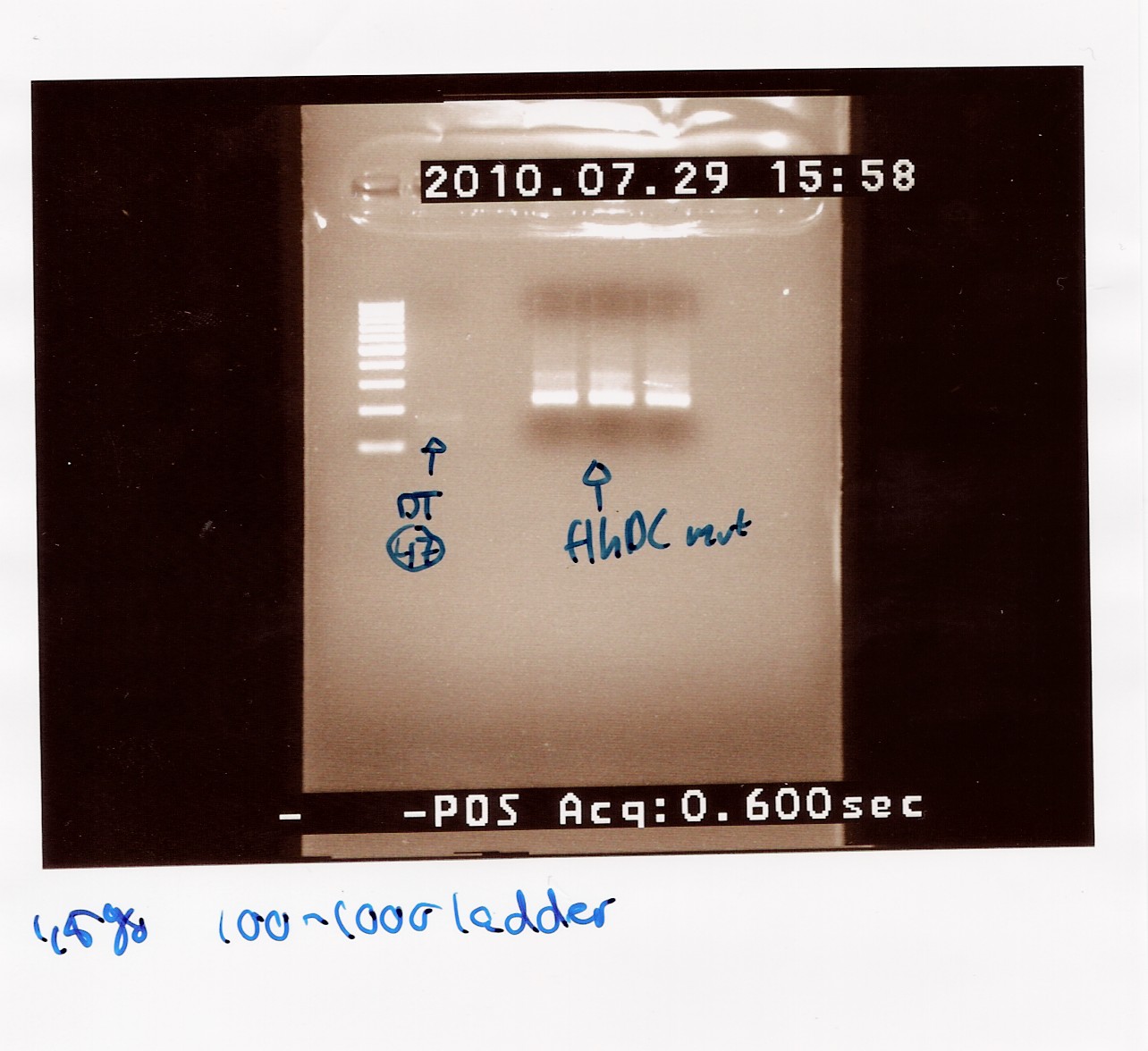
The picture shows a clear band at 200bp and smear. The FlhDC gene of 986bp is not seen on the gel.
--Louch07 07:26, 5 August 2010 (UTC)
Checking if Pst1 site in FlhDC mut is not pressent
Done by: Louise
Date: july 30th
Protocol:RD1.1
Notes: Two restrictions were made, one with native flhDC containing the Pst1 site (Freeze tube 13) and one with FlhDC mut which should not have a Pst1 site (freeze tube 49).
Restriction mixture:
13 ul water
1 ul Pst1 enzyme
2 ul FD green Buffer
5 ul PCR product
Results:
The result of this Pst1 test was very pour, and is redone later with other samples.

--Louch07 07:26, 5 August 2010 (UTC)
The gel was 1.5% and a 100-1000 ladder was used.
Incertion of flhD/C (mutated gene sequence) in pSB3K3 and pSB1C3
Done by: Maria and Sheila
Parts used: K343000 (flhD/C, mutated gene sequence), pSB3K3 and pSB1C3
==== Digestion and gelextraction ====
Protocol: [RD1.1]DE1.3
Notes: purified flhD/C and miniprep of pSB1C3 and pSB3K3 were digested using the EcoRI and PstI restriction enzymes.
Restriction mixture (FlhD/C):
| H2O | 38uL |
| FD green buffer | 8uL |
| EcoRI | 4uL |
| PstI | 4uL |
| FlhD/C DNA (white 48) | 30uL |
Restriction mixture (plasmid):
| H2O | 24uL |
| FD green buffer | 4uL |
| EcoRI | 2uL |
| PstI | 2uL |
| pSB3K3 or pSB1C3 | 10uL |
The digested samples were loaded onto a 1.5% agarose gel. Undigested FlhD/C and vector was used as controls. Hyperladder II and generuler DNA ladder mix (red) were used as markers.
Digested flhD/C is app. 980bp
pSB3K3 digested with EcoRI and PstI is 2713bp
pSB1C3 digested with EcoRI and PstI is 2035bp
DNA was extracted from gel according to protocol
Results:
==== Ligation ====
Protocol: LG1.2
Notes:
flhD/C is ligated with either pSB1C3 or pSB3K3.
3 ligation mixtures with ratios of 1:3, 1:6 and 1:9 (vector:flhD/C)was prepared.
20-30ng of vector was used for each ligation.
Ligation mixtures (FlhD/C in pSB3K3):
| Lig. 1(1:3) | Lig. 2 (1:6) | Lig. 3 (1:9) | |
| T4 ligase bf. | 2uL | 2uL | 2uL |
| T4 ligase | 1uL | 1uL | 1uL |
| pSB3K3 | 2.5uL | 2.5uL | 2.5uL |
| FlhD/C | 2uL | 4.5uL | 6.5uL |
| H20 | 12.5uL | 10uL | 8uL |
Ligation mixtures (flhD/C in pSB1C3):
| Lig. 1(1:3) | Lig. 2 (1:6) | Lig. 3 (1:9) | |
| T4 ligase bf. | 2uL | 2uL | 2uL |
| T4 ligase | 1uL | 1uL | 1uL |
| pSB1C3 | 2uL | 2uL | 2uL |
| FlhD/C | 4uL | 5uL | 6uL |
| H20 | 11uL | 10uL | 9uL |
All samples were incubated at 17 degrees until they were used for transformation.
Follow-up colony PCR
Date: 26/7
Methods: Colony PCR
Protocol: CP1.3
Experiment done by: Maria, LC
Notes: We made a sample for every plate from the last colony PCR. Out of the 29 samples we could only run 25 at once in the PCR machine. Plate 23 was missing, so there is no sample for it.
Results: 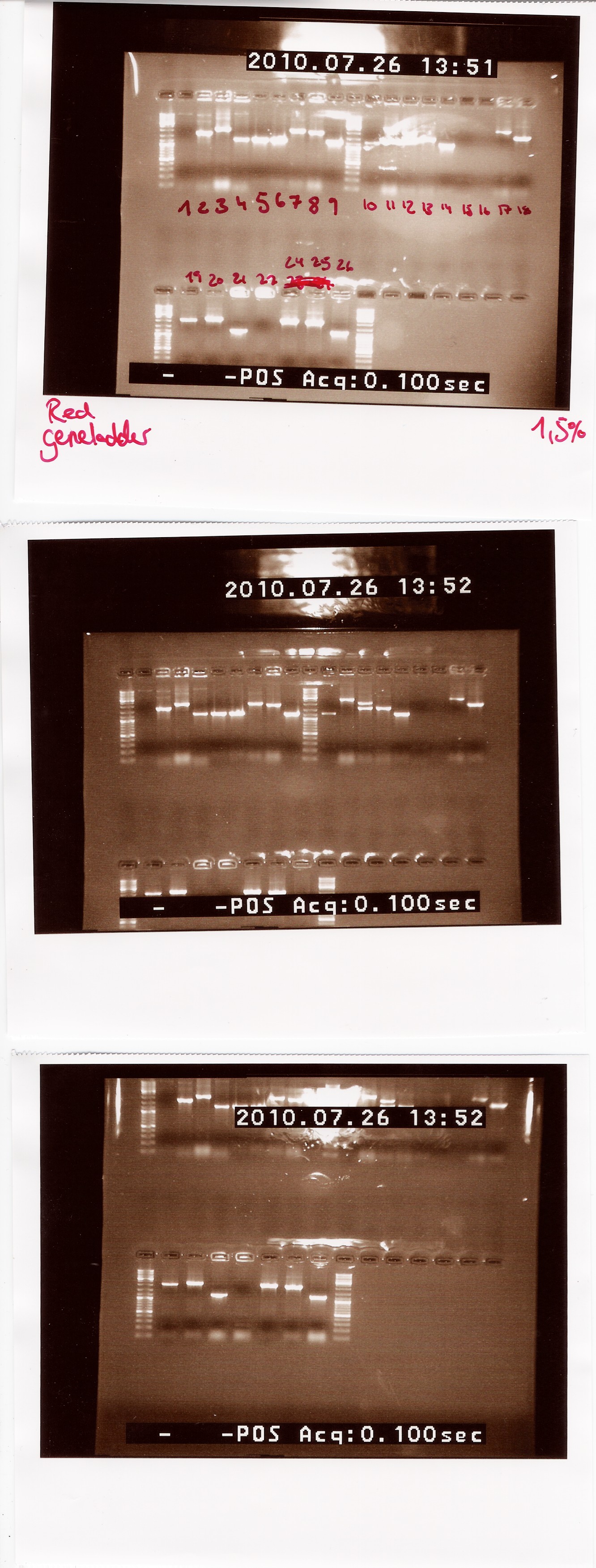
Lengths of PCR products:
1200: 4, 5, 6, 9, 10, 14, 21, 26
1500: 13
1750: 2
1900: 8, 25
2000: 3, 7, 12, 18, 19, 24
2200: 20
2500: 11, 17
Colonies 1, 15, 16 and 22 gave no result.
Analysis: Since we had so many different lengths, we will cut one from each length, specifically colony: 2, 8, 11, 13, 20, 24, 26.
Restriction digest of PCR product from 26/7
Date: 26/7
Methods: Restriction digest
Protocol: RD1.1
Experiment done by: Maria, LC
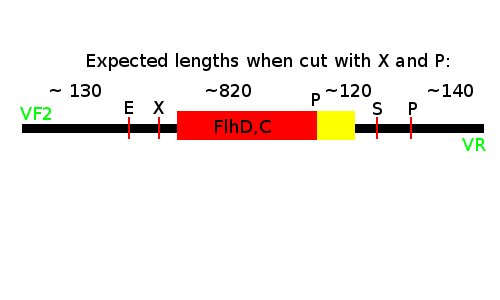
Notes: We took one of each length PCR product and cut them with XbaI and PstI. The especially interesting one was sample 26, since we suspected it to be a succesful ligation. If it is FlhD,C we would expect it to get cut like this:
We loaded the cut and uncut sample next to each other. They got loaded as follows and RFP was used as a control:
| Lane |
Sample |
Cut/Uncut |
| 1 |
2 |
c |
| 2 |
2 |
u |
| 3 |
8 |
c |
| 4 |
8 |
u |
| 5 |
11 |
c |
| 6 |
11 |
u |
| 7 |
13 |
c |
| 8 |
20 (loading error) |
u |
| 9 |
20 |
c |
| 10 |
13 (loading error) |
u |
| 11 |
24 |
c |
| 12 |
24 |
u |
| 13 |
26 |
c |
| 14 |
26 |
u |
| 15 |
7(RFP) |
c |
| 16 |
7(RFP) |
u |
Results: 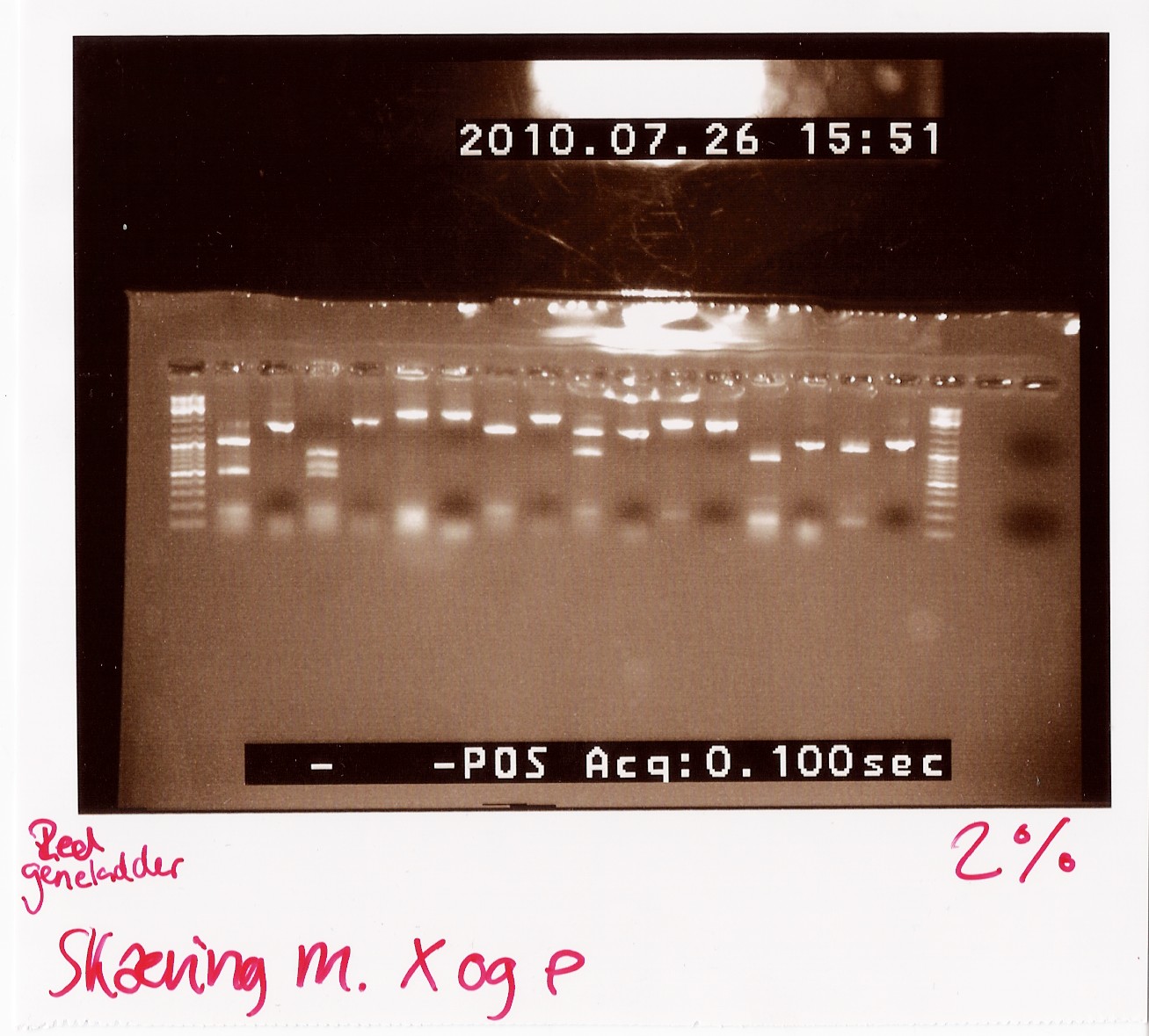
Analysis: Sample 26 looks very close to the right lengths, so we will prepare some of the 1200 BP length PCR products for sequencing.
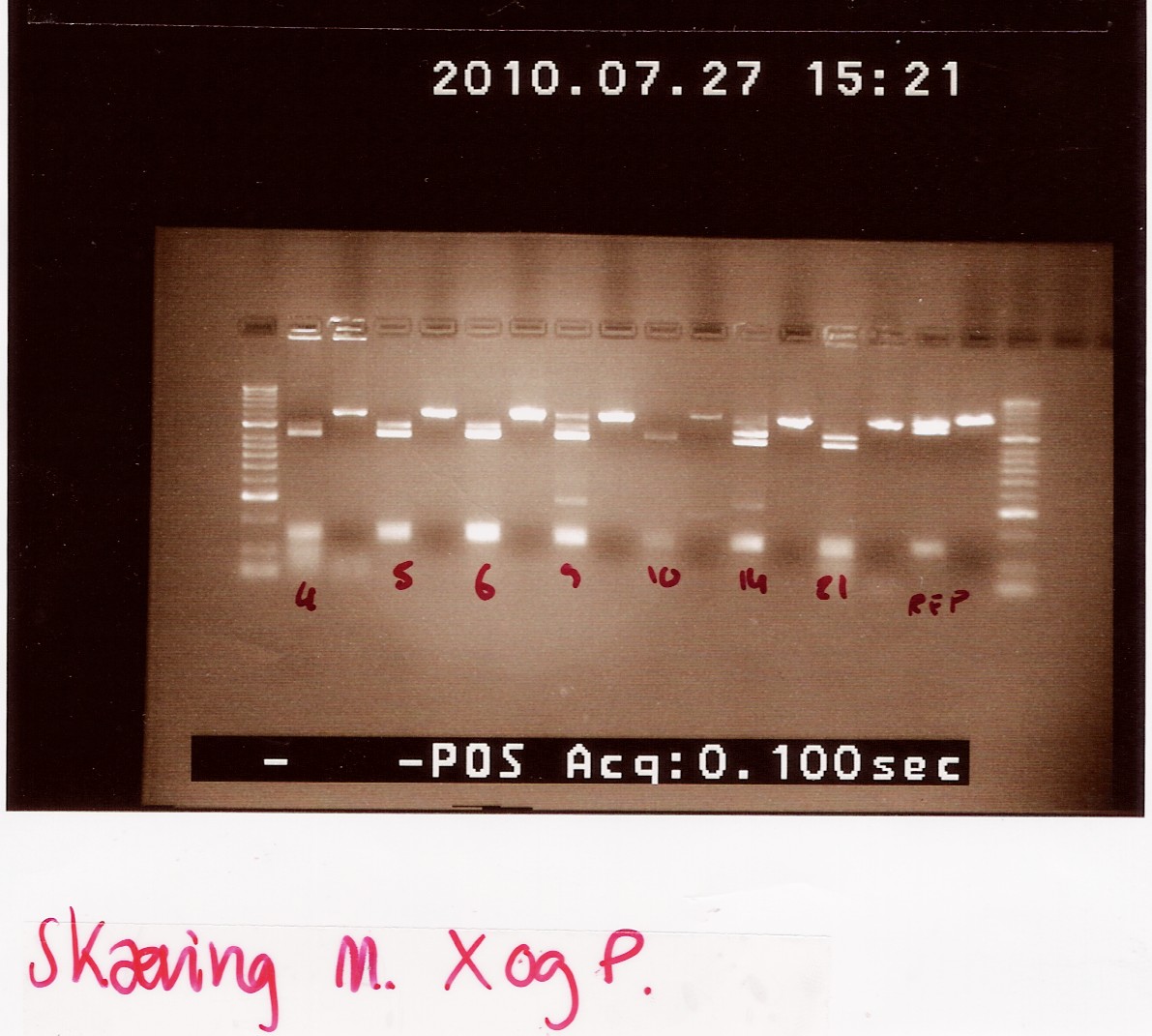
Miniprep of follow-up colony PCR
Date: 27/7
Methods: ON, Miniprep
Protocol: MP1.1
Experiment done by: LC
Notes: Samples L4, L5, L6, L9, L10, L14, L21 and L26 (L for ligation from the follow-up colony PCR) miniprepped and NinaB samples from the 4 frozen cultures of it.
Results: 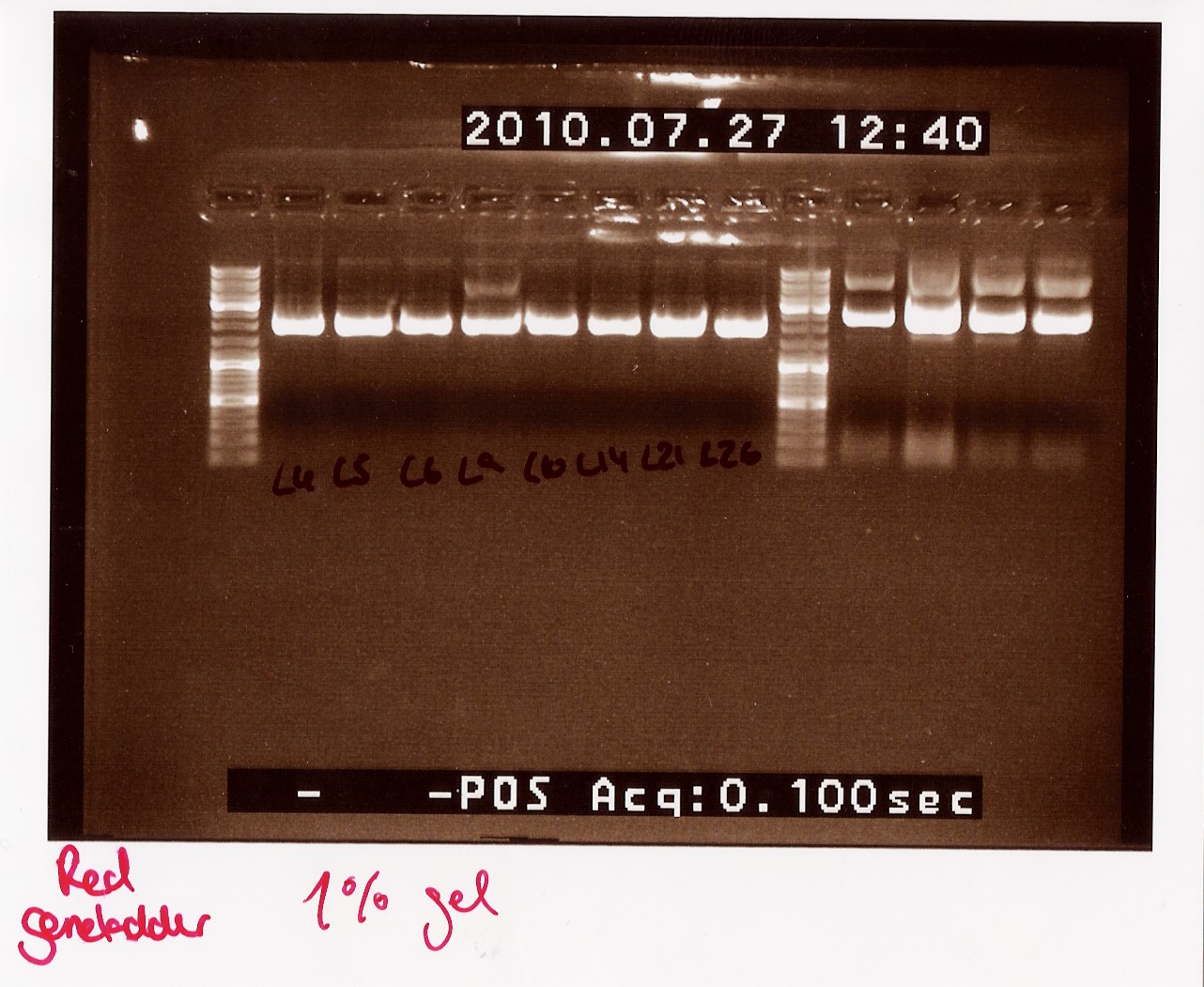
All ligations were around the same length, just under 2000 BP. Even though the correct length should have been about 1000 BP longer, this is not surprising since uncut plasmids often show 1000 BP less in length. NinaB was also too short and there was a second band showing higher up in the gel (weird!). The concentrations averaged around 80ng/ul for the samples.
Analysis: We decided to do a new miniprep where we start with a higher concentrated overnight culture, which we will achieve through boosting the ON before doing the miniprep.
Restriction digest of PCR product from 26/7
start date: 27/7
Methods: Restriction digest, gel electrophoresis
Protocol:RD1.1
Experiment done by: Maria, LC
Notes:
All colonies with PCR products of 1200bp (see Follow up coloni PCR) was digested with XbaI and PstI.
Coloni #4,5,6,9,10,14 and 21 was selected.
Digested RFP PCR product (#6 from Coloni PCR) was used as controle.
If it is FlhD,C we would expect it to get cut like this:

Digested samples were loaded along with undigested ones. Samples were loaded onto a 2% gel. Hyper ladder 2 was used as marker.
Loading scheme:
| Lane |
Sample |
Cut/Uncut |
| 1 |
4 |
c |
| 2 |
4 |
u |
| 3 |
5 |
c |
| 4 |
5 |
u |
| 5 |
6 |
c |
| 6 |
6 |
u |
| 7 |
9 |
c |
| 8 |
9 |
u |
| 9 |
10 |
c |
| 10 |
10 (loading error) |
u |
| 11 |
14 |
c |
| 12 |
14 |
u |
| 13 |
21 |
c |
| 14 |
21 |
u |
| 15 |
6(RFP) |
c |
| 16 |
6(RFP) |
u |
Results:
gel electrophoresis:
Analysis:
The lanes containing digested PCR product from coloni #5, 6 and 21 have a band smear around 100-200bp,and a band at around 800bp.
This may indicate correct inserted flhD/C.
ON cultures are made of each of these colonies to use for boost Miniprep.
--Tipi 13:26, 28 July 2010 (UTC)
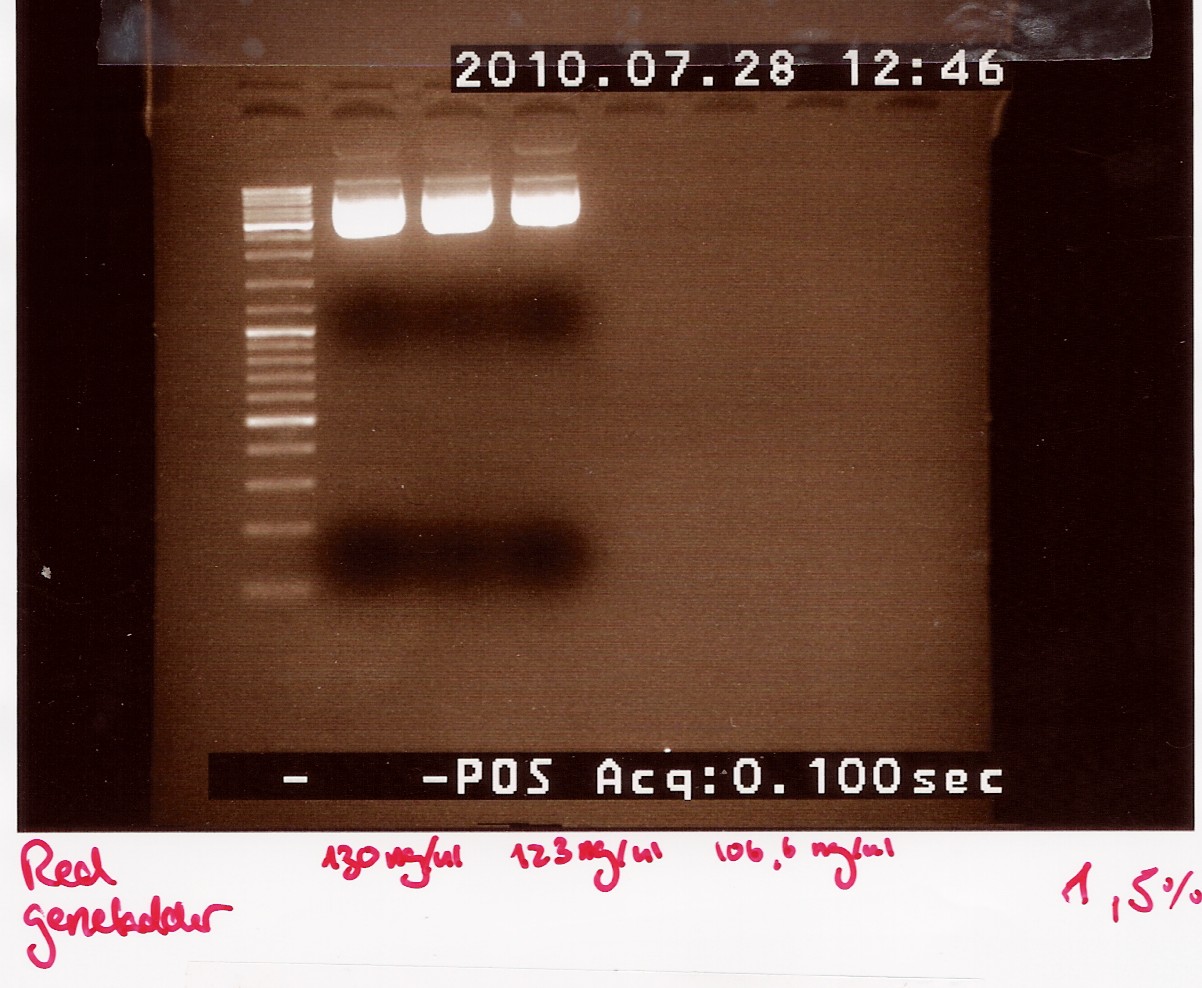
Miniprep of ligation plasmids
start date: 27/7
Methods: miniprep, gel electrophoresis, nanodrop
Protocol:MP1.3
Experiment done by: LC
Notes:
Miniprep was made of ON cultures of coloni #5,6 and 21 (see Follow up coloni PCR)
Samples were loaded onto a 1.5% agarose gel. Gener ruler DNA ladder mix was used as marker.
Loading scheme:
| Lane |
sample |
| 1 |
#5 |
| 2 |
#6 |
| 3 |
#21 |
Results:
Gel electrophoresis:
Analysis:
we have purified the plasmids.The concentrations are high but not as high as expected when the cells are boosted prior to the miniprep.
In order to obtain an even higher concentration transfer all 5mL ON culture to 20mL pre heated LB media. run all 25mL new culture as 1 miniprep.
Due to the low concentrations we need to dry down our samples before they are ready for sequentation.
--Tipi 14:05, 28 July 2010 (UTC)
Digestion and gel extraction of pSB3C5 and J13002
Done by: Pernille
Date: the 26th of july
Protocol: [RD1.1] and gelpurificaton (DE1.3)
Notes:because the concentration for both plasmid and biobrick is rather small I dobbelt the volumen of the reagent compared to the protocol. in the soultion contaning the brick the volumen of the PCR product was triple and to adjust the total volumen i added 5ul less water. We made 2 Restriction mixtures which both were 4 times the mixture in the protocol. both the plasmid and brick was cut at the E and P site. After the cutting the plasmid and brick was run on seperate gels because of there different size. the plasmid on a 1,5% gel and brick on a 2% agarose gel. When I observed the gel under the UV lamp the was no visible band on the gel were the brick was loaded. therefore it was only possible to do DNA extrated from the gel contraining the plasmid. The Dna was eluted in 10ul of water and I eluted two times. The DNA concentration was measured on the NaonDrop and was found to 55,41ng/ul for the first elution and 28,03ng/ul for the second. the to samples are pooled and stored in a freeze tube.
Group: Retinal
Colony PCR and extraction of ninaB gene from transformants
Start date: 24/7
Methods: colony pcr, fermentas fast digest
Protocols: CP1.3, RD1.1
Colony PCR on transformants using ninaB fwd and rw primers
Date: 24/7
Done by: Christian
Methods: Colony PCR
protocos: CP1.3
Primers were prepared by ellution to 100uM for storage, and again by a factor 10 for the reaction. 10 wells were loaded, and their colonies were streaked.
Annealing temperature set to 55°C and elongation set to 2 minutes, otherwise standard taq protocol was followed.
Results:_Expected length at 1920 BP did not show. In fact no bands showed, except somethin like a primer smear around 100bp.
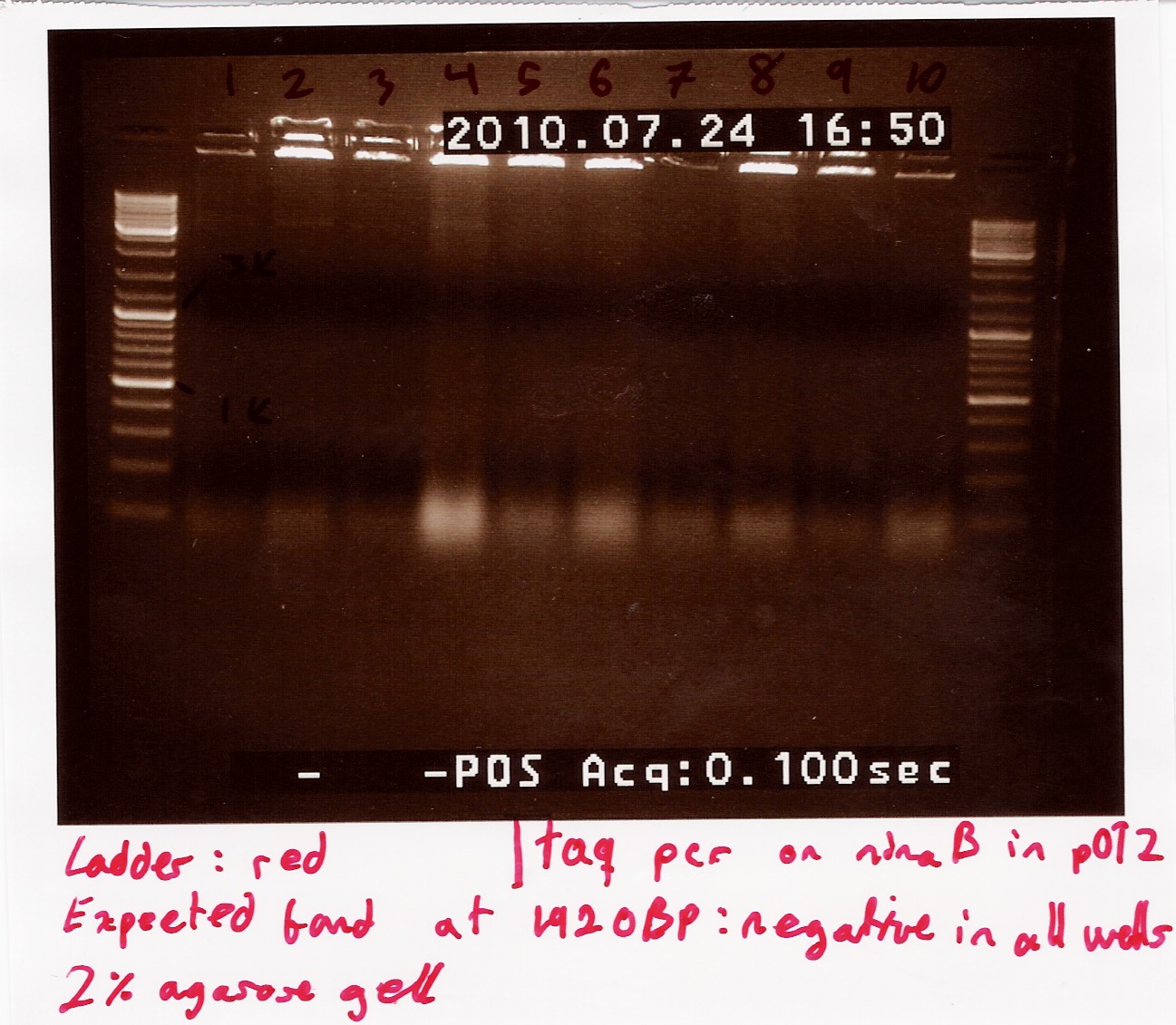
Analysis: Anealing temperature might have been set to high, as the primers were constructed to aneal at 55°. another run will be done on miniprepped plasmids, with a different program.
Restriction digest on miniprepped pOT2 plasmids with ninaB insert using EcoRI
Date: 27/7
Done by: Christian
Methods: Restriction digest
protocos: RD1.1
A restriction digest was carried out to determine the length of the pOT2 plasmid and insert. protocol was followed to point, using miniprep as template dna.
Results: strange things going on at different lengths, and nothing matches the expected length of about 3800BP.
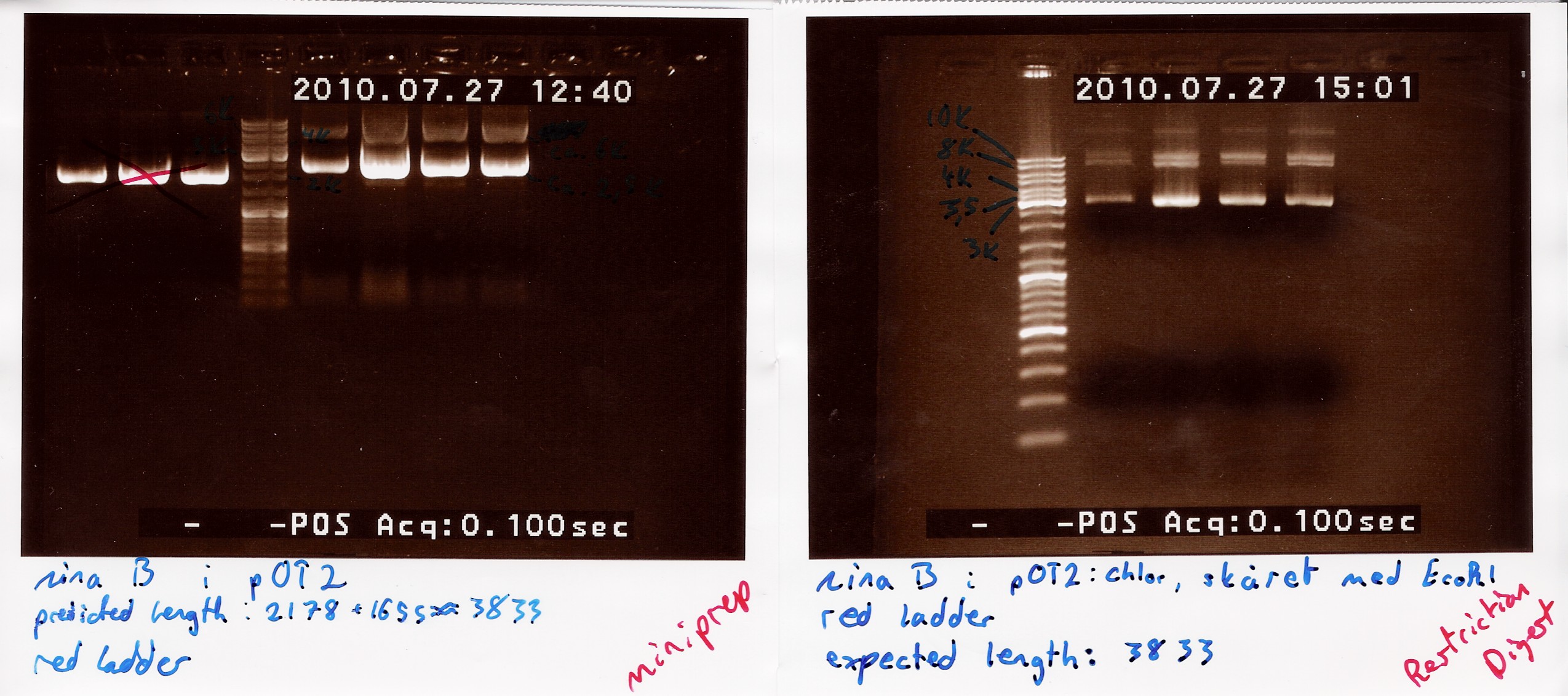
Analysis: Another digest is needed to show that plasmid length is wrong, as it seems unlikely with comercial plasmids.
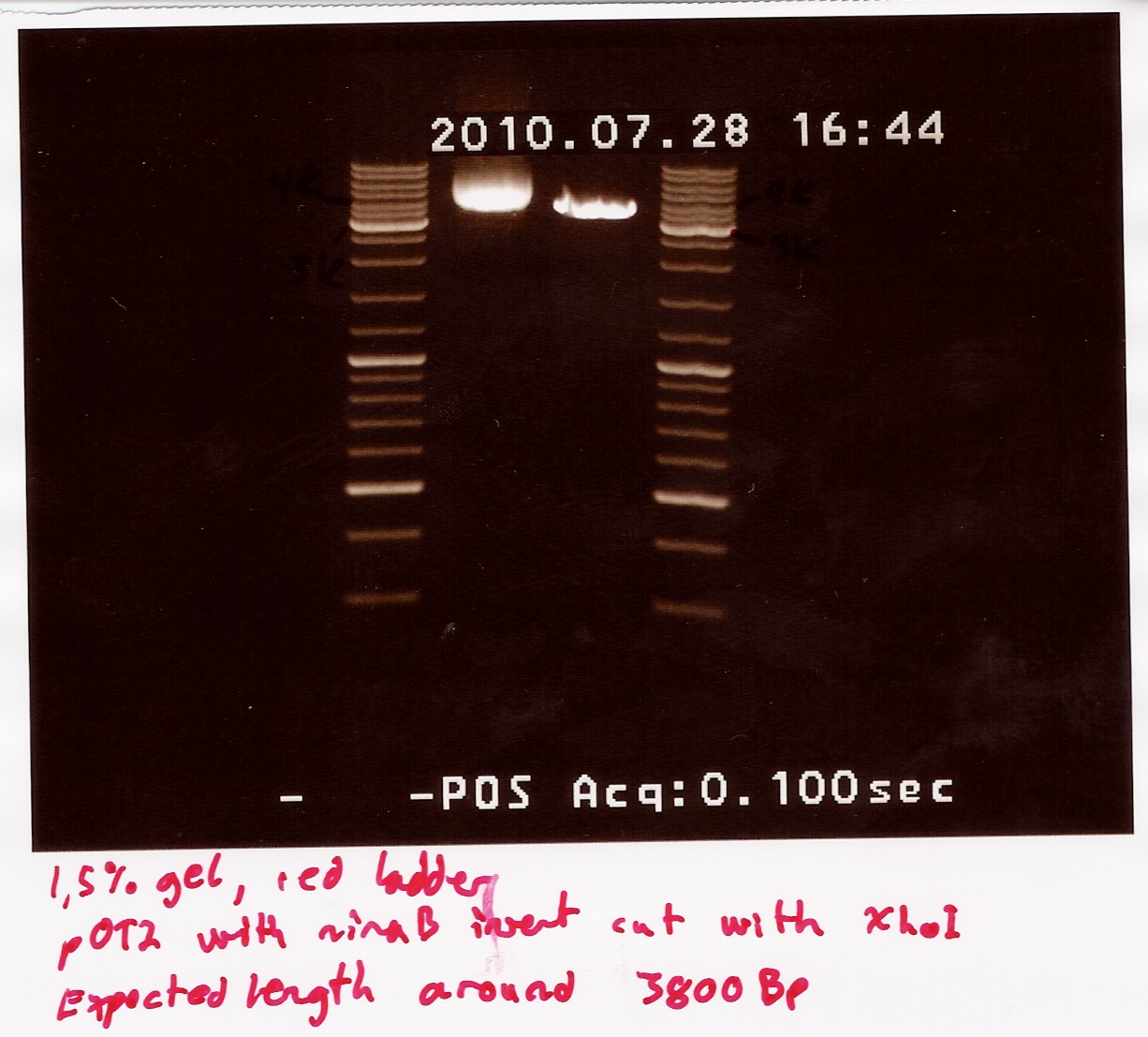
Restriction digest on miniprepped pOT2 plasmids with ninaB insert using EcoRI and XhoI
Date: 28/7
Done by: Christian
Methods: Restriction digest
protocos: RD1.1
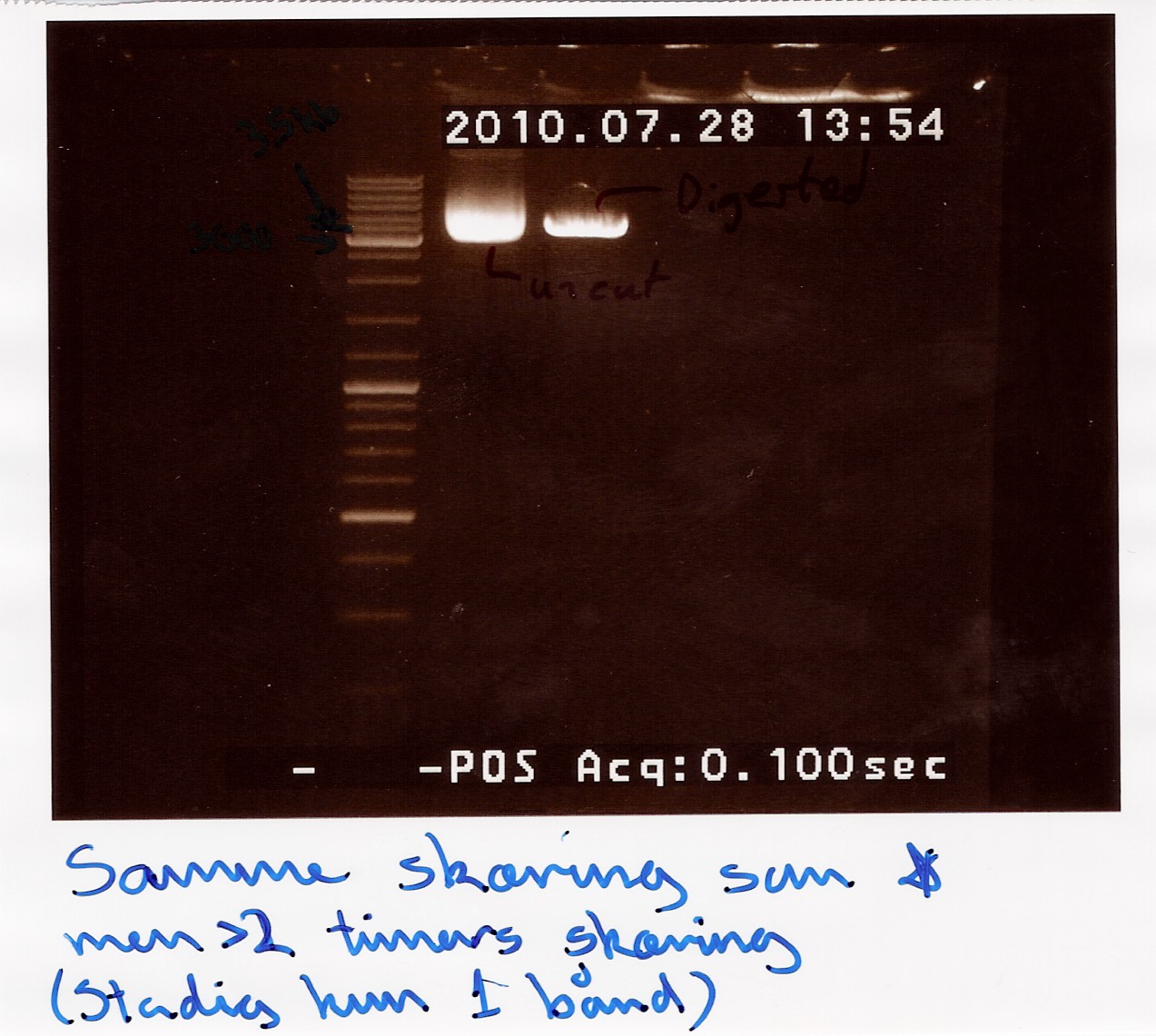
A restriction digest was carried out to determine the length of the pOT2 plasmid and insert. protocol was followed to point, using miniprep as template dna. Both XhoI and EcoRI were used, as these were the enyzmes originally used to create insert and cut plasmid for ligation according to fruitfly under the GH section, which our pOT2 plasmids falls under. protocol was followed to point, except for the digest running for 2 hours.
results: Digest showed a band close to the 3.8kb marker expected for the full length plasmid, concurrent with only one digest working. (although an earlier digest with EcoRI alone showed a different band.) Miniprepped plasmid had also "moved up" the ladder
analysis: Something fishy is up with this plasmid, and the digest. one more will be run only using XhoI.
Restriction digest on miniprepped pOT2 plasmids with ninaB insert using EcoRI and XhoI
Date: 28/7
Done by: Christian
Methods: Restriction digest
protocos: RD1.1
A restriction digest was carried out to determine the length of the pOT2 plasmid and insert. protocol was followed to point, using miniprep as template dna. XhoI was used as cutting enzyme, and digest was allowed to run for 30 mins.There were problems with the first gells, so a second gel was run.
results: Gel showed a band between 3.5K and 4k markers, consistent with earlier results, indicating that the plasmid and insert have correct length. Strangely the cut segment has traveled further through the gell than the un-cut control.
analysis: There realy is something fishy going on, or as we say in Denmark, there are owls in the marsh. At least the plasmid seems correct.
Ligation of J13002 promotor rbs into pSB3T5 assembly plasmid.
Start date: 26/7
Methods: Restriction Digest, Gel Extraction, Ligation, Transformation (not done by me) and colony pcr
Protocols: CP1.3, LG1.3, RD1.1, DE1.3
Restriction digest, Gel extracton and ligation of BBa_J13002 and pSB1C3
Date: 27/7
Done by: Christian
Methods: Restriction digest, Gel extraction, ligation and transformation (not by me)
protocos: RD1.1, DE1.3, LG1.3
Restriction digest using EcoRI and PstI and gel extraction were done following protocol. at beginning of ligation concentrations were: 3.7ng/ul part at 95BP and 8ng/ul plasmid at 3215BP. four tubes were run, 1 at 3:1 ration, two at 6:1 and one at 9:1. iGEM consensus protocol was followed, with 20ng vector as starting point. Tubes were cooled for transformations in the morning.
Results: Quite low concentrations were reached, but the iGEM protocol should ensure the same molar ratio and total concentration as was planned for.
Analysis: Everything is going according to plan for now.
colony PCR using taq on ligated BBa_J13002 in pSB1C3
Date: 28/7
Done by: Christian
Methods: colony pcr
protocos: CP1.3
Colony pcr was run on 8 colonies, and plates were streaked and incubated at 37° ON. protocol was followed to point, with enzyme added to mix. PErhaps too little colony was scraped of original plates, and some tubes were defective, letting out steam during pcr.
Results: A band showed at around 300bp in well 4.1. The expected length was 312bp. wells3.3 and 3.4 show a band outside the blue ladder, that could be rfp (since this was the original insert.) tube 3.2 was ruined.

Analysis: Well 3.2 shows promising results. ON and miniprep will be made, and it will be sent for sequencing.
Group: Photosensor
Amplification of BOO15 in pSB1AK3 and J13002 in pSB1A2
Date: 27/7
Done by: Maria
Methods: pfu PCR, gel electrophoresis
protocos: CP1.1
Notes:
gel purified PCR product of BOO15 and J12003 (25 and 26 white) was used as template.
Premix:
| Template |
2uL |
| pfu buffer + MgSO4 |
40uL |
| dNTP's |
12uL |
| VF2 |
12uL |
| VR |
12uL |
| H20 |
304uL |
| pfu polymerase |
3uL |
PCR tubes and premix were kept at ice at all times. pfu buffer was added directly to the premix, before it was distributed into the PCR tubes.
pfu program:
| start |
94C |
3min |
| denaturating |
94C |
2min |
| annealing |
55C |
30s |
| elongation |
72C |
30s |
| go to |
2 |
rep. 29x |
| end |
72C |
2min |
| hold |
4C |
PCR samples was loaded onto a 2% agarose gel. Gene ruler 100bp DNA ladder (blue) was used as marker.
Loading scheme:
| Lane |
1 |
2 |
3 |
4 |
| J13002 |
J13002 |
B0015 |
B0015 |
Results:
gel electrophoresis:
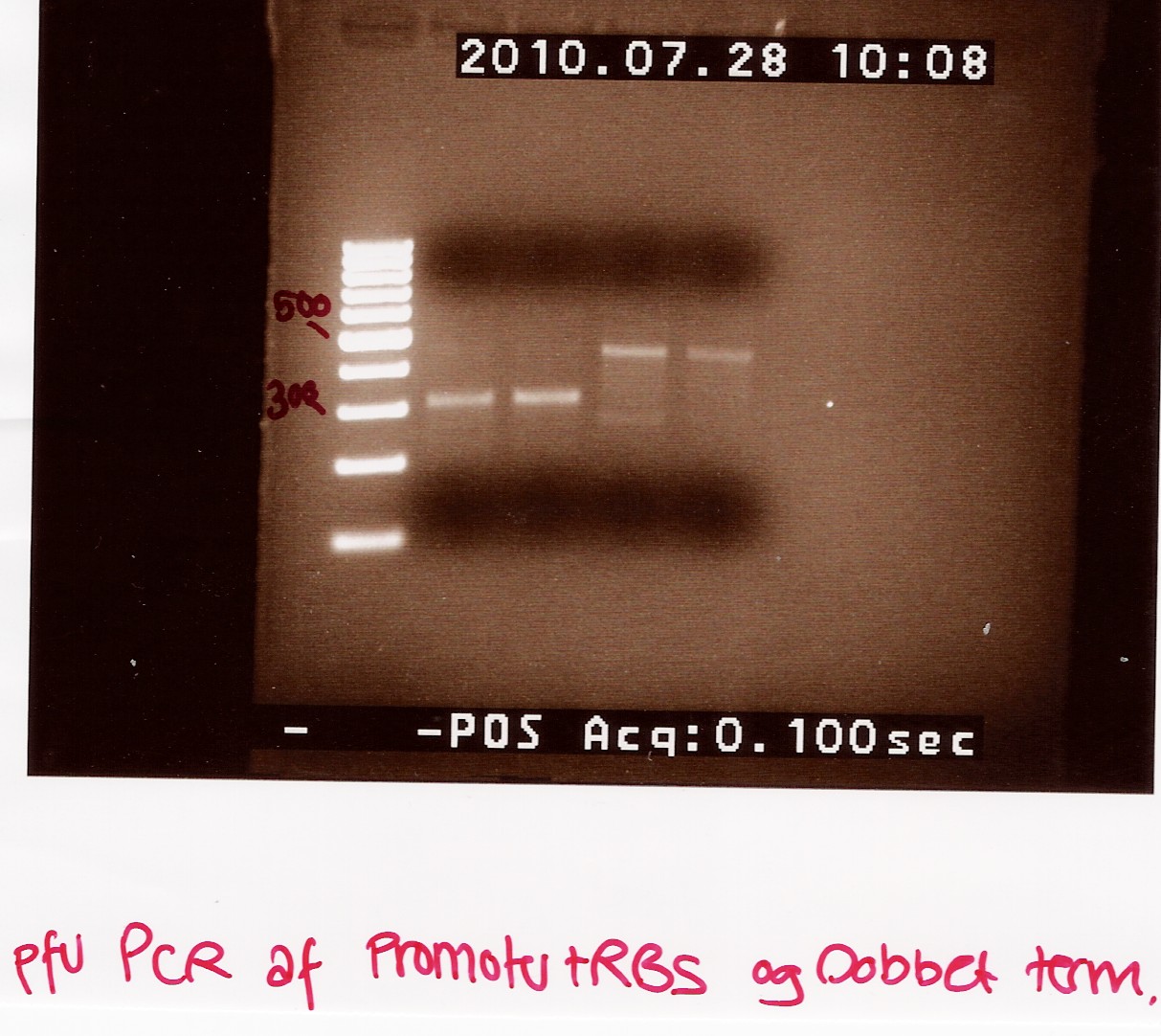
Analysis:
The PCR product is OK and is used as template for pfu PCR.
Surplus PCR product is pooled and stored at -20degrees.
J13002 is stored as 42 (white)
BOO15 is stored as 43 (white)
--Tipi 06:52, 29 July 2010 (UTC)
Coloni PCR of photosensor in pUC19 from transformation 27/7
Date: 28/7
Done by: Pernille
Methods: Taq PCR, gel electrophoresis
protocos: CP1.3
Notes:
6 colonies from transformation 27/7 is selected and used for coloni PCR.
Premix:
| 10x taq buffer |
20uL |
| MgCl2 |
8uL |
| VF2 |
8uL |
| VR |
8uL |
| dNTP's |
4uL |
| H20 |
28uL |
| taq polymerase |
3uL |
PCR tubes are marked PS1.A-F
Taq PCR program:
| start |
94C |
2min |
| denaturating |
94C |
1min |
| annealing |
55C |
1min |
| elongation |
72C |
3min |
| go to |
2 |
29x |
| end |
72C |
3min |
| hold |
4C |
sizes:
Photosensor: 1914bp
VF2-VR (without insert): 845bp
VF2-VR (with Photosensor as insert): 2759bp
The PCR samples was loaded onto a 1% agarose gel. Gene ruler DNA ladder mix (red) was used as marker.
Results:
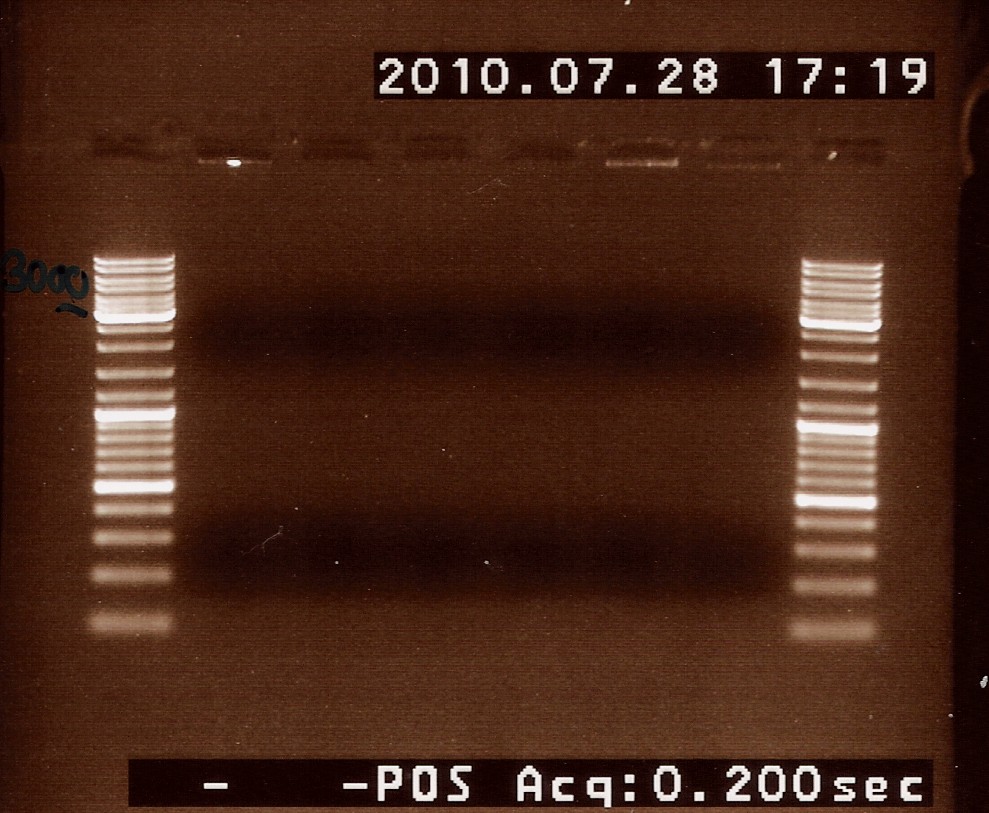
Analysis:
No bands are seen in any of the samples. This could be because the VF2 and VR primers does not anneal to the plasmid.
Custom primeres must be ordered.
ON cultures are made of some of the colonies to use for freeze cultures.
--Tipi 07:18, 29 July 2010 (UTC)
Taq PCR of photosensor in pUC19
Date: 29/7
Done by: Maria
Methods: Taq PCR, gel electrophoresis
protocos: CP1.3
Notes:
To ensure that the photosensor is inserted in pUC19 a taq PCR using the DNA (tube 40 white) as template and VF2 and VR as primers.
1uL template is used pr. PCR reaktion (2 PCR reaction are prepared)
Premix:
| 10x taq buffer |
7.5uL |
| MgCl2 |
3uL |
| VF2 |
3uL |
| VR |
3uL |
| dNTP's |
1.5uL |
| H20 |
52.5uL |
| taq polymerase |
1uL |
H2O normally used to lyse the colonies are used in the premix.
PCR tubes are marked PS2.A-B
Taq PCR program:
| start |
94C |
2min |
| denaturating |
94C |
1min |
| annealing |
55C |
1min |
| elongation |
72C |
3min |
| go to |
2 |
29x |
| end |
72C |
3min |
| hold |
4C |
PCR samples are loaded onto a 1% agarose gel. Gene ruler DNA ladder mix (red) is used as marker.
Results:
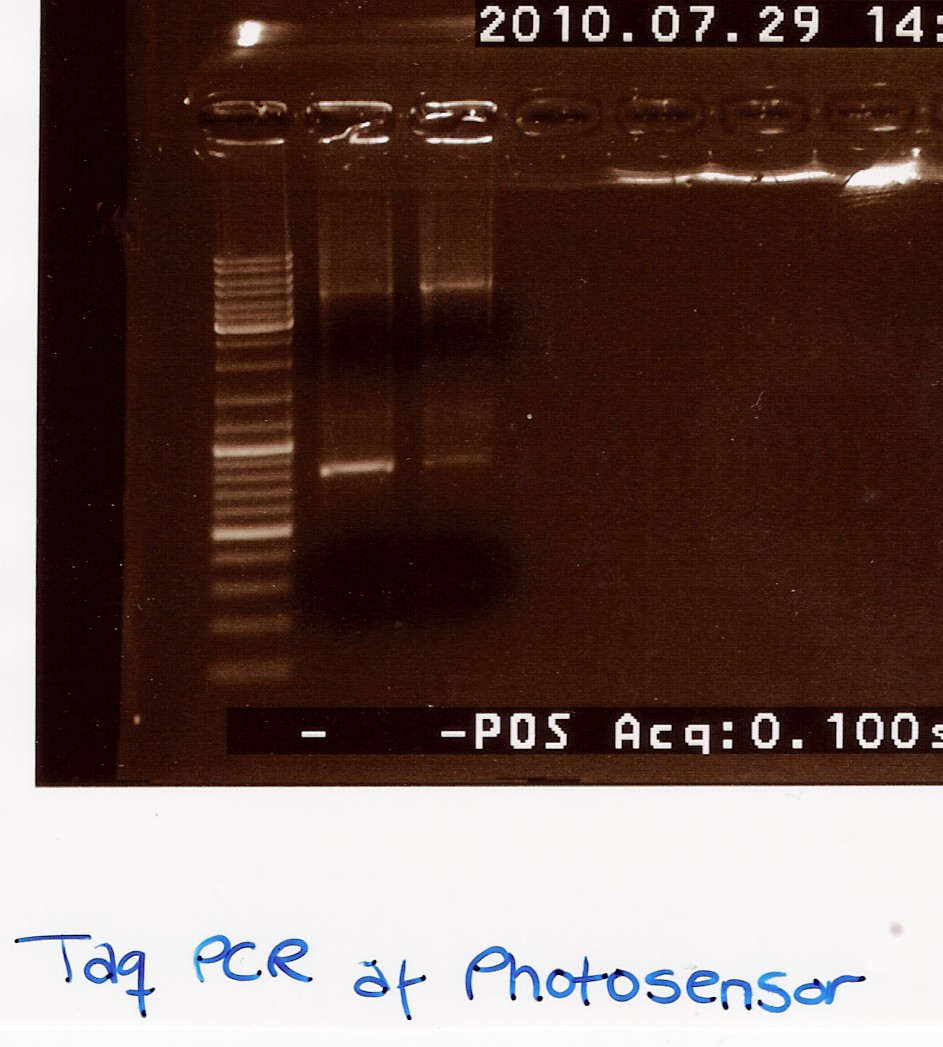
Analysis:
Two weak bands at app. 4500bp and 850bp respectively. The upper band could correspond to the pKJ606 plasmid with insert, and the lower band might indicate VF2-VR without insert, rendering these results rather inconlclusive. Therefore an ON culture for miniprep was made.
--Lclund 06:47, 4 August 2010 (UTC)
Restriction digest and PCR on ninaB plasmids
Date: 29/7
Done by: Christian and Marie
Methods: Restriction igestion, gel electrophoresis, gradient Taq PCR
protocos: ??
Notes:
Three restriction reactions were made:
1: pOT2 with ninaB cut with EcoRI (E)
2: pOT2 with ninaB cut with XhoI (X)
3: pOT2 with ninaB cut with EcoRI and XhoI (EX)
A 1.5% agarose gel was run:
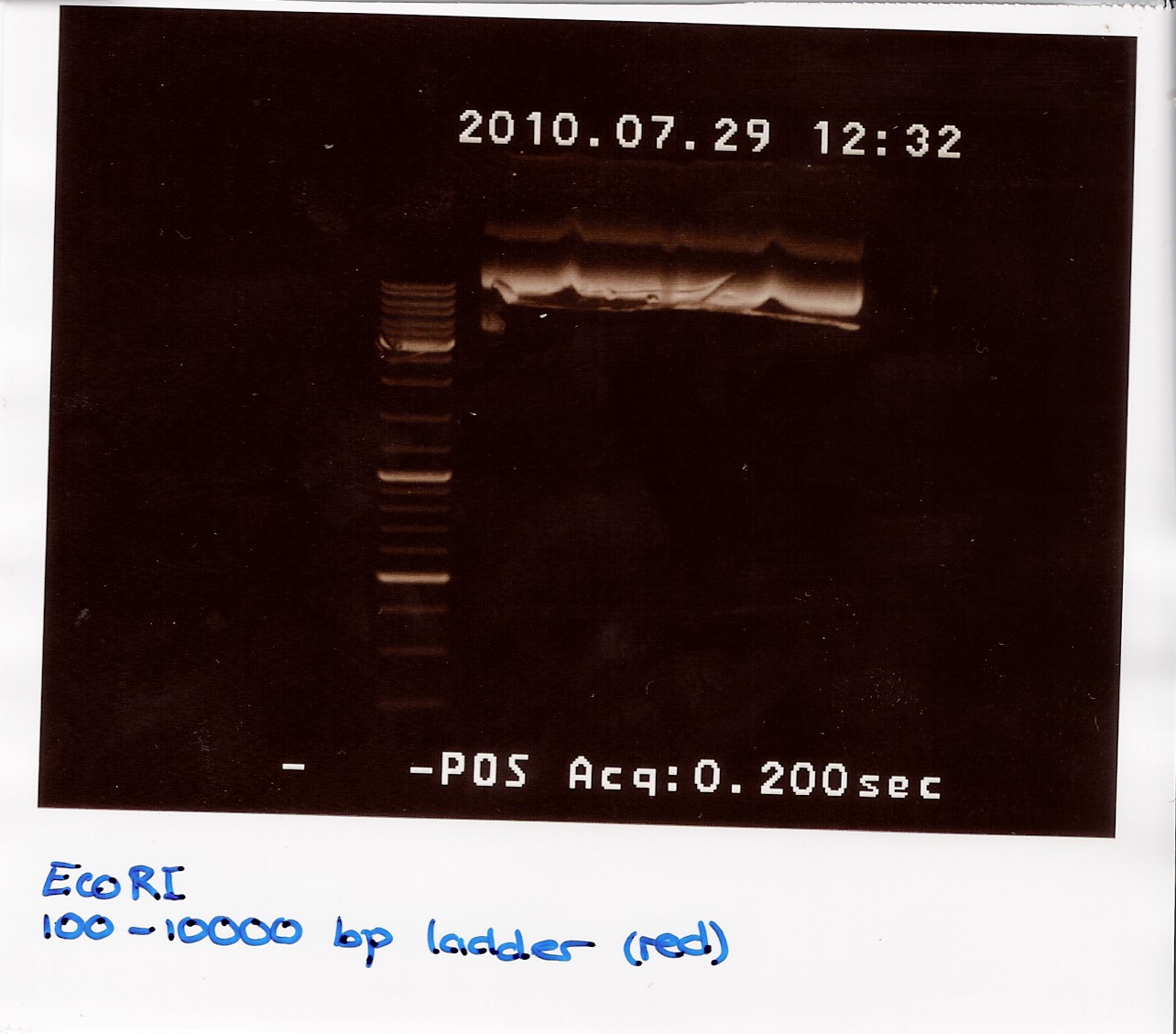
Nucleic acid concentrations were measured with NanoDrop after gel extraction and purification:
Gradient PCR was run at the following temperatures:
| Sample no. |
PCR column |
Temperature |
| 1 |
2 |
45.3 °C |
| 2 |
4 |
48.5 °C |
| 3 |
6 |
53.4 °C |
| 4 |
8 |
58.7 °C |
| 5 |
10 |
63.1 °C |
| 6 |
12 |
65.1 °C |
 "
"