Team:SDU-Denmark/K343006
From 2010.igem.org
(Difference between revisions)
m (→HPLC determination of beta-carotene and retinal production) |
(→UV-Vis determination of beta-carotene and retinal production) |
||
| (2 intermediate revisions not shown) | |||
| Line 16: | Line 16: | ||
MG1655-pSB1A2-K274210/pSB1C3-K343005 (double transformants)<br> | MG1655-pSB1A2-K274210/pSB1C3-K343005 (double transformants)<br> | ||
Both K343005 and K274210 were constitutively expressed.<br><br> | Both K343005 and K274210 were constitutively expressed.<br><br> | ||
| - | Since the first experiment showed no or small amounts of product in cells grown to exponential phase the measurements were performed on cells incubated for 20 hours at 37 degrees Celcius | + | Since the first experiment showed no or small amounts of product in cells grown to exponential phase the measurements were performed on cells incubated for 20 hours at 37 degrees Celcius. The graphs obtained from the experiment are presented below:<br> <br> |
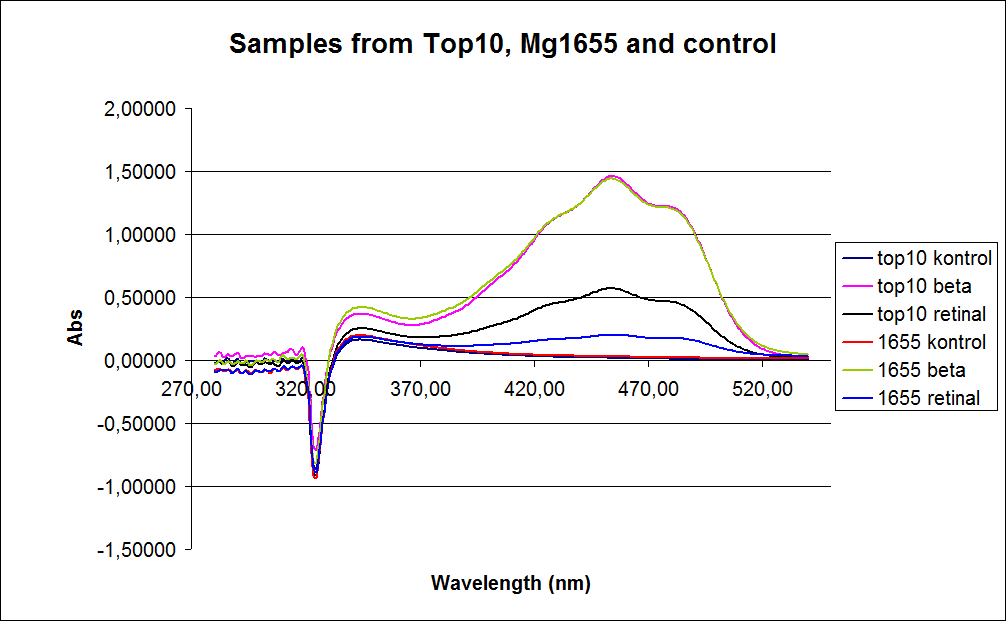
| - | [[Image: Team-SDU-denmark-Samples form top10 mg1655 and control.png |thumb|400px|center|UV-VIS spectre of six ''E. coli'' strains: Wild type TOP10, Wild type MG1655, TOP10-pSB1A2-K274210, MG1655-pSB1A2-K274210, TOP10-pSB1A2-K274210/pSB1C3-K343005 (double transformants), MG1655-pSB1A2-K274210/pSB1C3-K343005 (double transformants)]] | + | [[Image: Team-SDU-denmark-Samples form top10 mg1655 and control.png |thumb|400px|center|'''Figure 1:''' UV-VIS spectre of six ''E. coli'' strains: Wild type TOP10, Wild type MG1655, TOP10-pSB1A2-K274210, MG1655-pSB1A2-K274210, TOP10-pSB1A2-K274210/pSB1C3-K343005 (double transformants), MG1655-pSB1A2-K274210/pSB1C3-K343005 (double transformants)]] |
<br><br> | <br><br> | ||
In the spectrum a sudden drop between 330 and 320 nm occurs, this can be caused when the samples was auto zeroed according to acetone which is the solvent used in the extraction of the beta-carotene and Retinal.<br> | In the spectrum a sudden drop between 330 and 320 nm occurs, this can be caused when the samples was auto zeroed according to acetone which is the solvent used in the extraction of the beta-carotene and Retinal.<br> | ||
The graph also shows that cell material interferes with the UV-vis measurement where the retinal has the strongest absorption. Therefore, an organic separation is required prior to the measurements.<br> | The graph also shows that cell material interferes with the UV-vis measurement where the retinal has the strongest absorption. Therefore, an organic separation is required prior to the measurements.<br> | ||
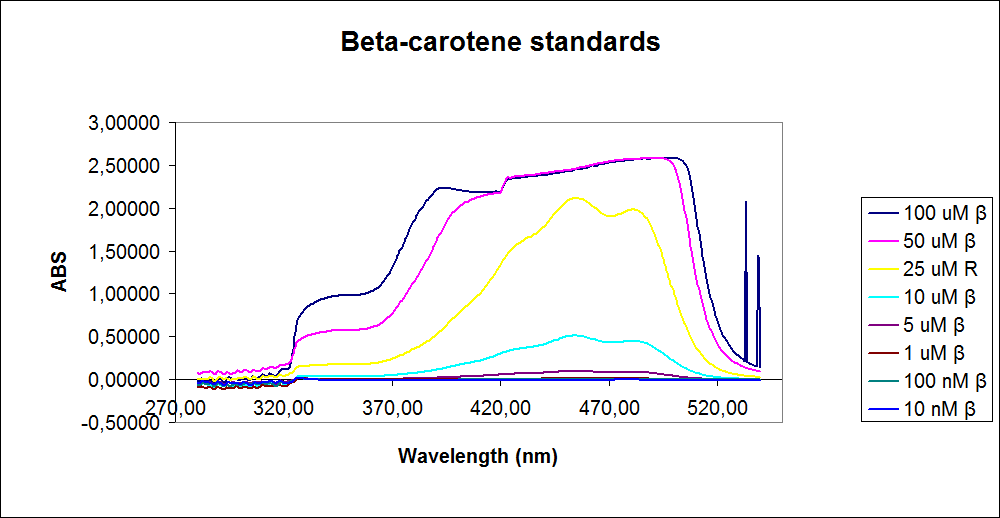
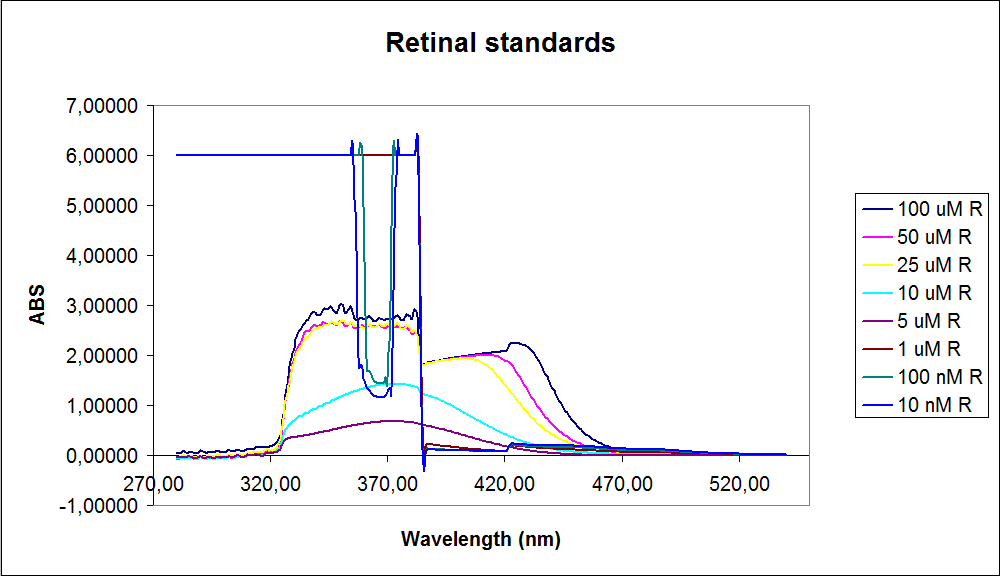
In order to assess the results, standard dilutions of beta-carotene and retinal were made and measured. The concentrations were 1mM, 100 µM, 50 µM, 25 µM, 10 µM, 5 µM, 1 µM, 100 nM and 10 nM. The standard dilutions were measured on the spectrophotometer. The resulting graphs are presented below:<br><br> | In order to assess the results, standard dilutions of beta-carotene and retinal were made and measured. The concentrations were 1mM, 100 µM, 50 µM, 25 µM, 10 µM, 5 µM, 1 µM, 100 nM and 10 nM. The standard dilutions were measured on the spectrophotometer. The resulting graphs are presented below:<br><br> | ||
| - | [[Image: Team-SDU-denmark-Beta-carotene standarts.png|thumb|350px|left|UV-VIS of beta-carotene at different concentrations. The spectre looks reliable]][[Image: Team-SDU-denmark-Retinal standarts.png|thumb|315px|center|UV-VIS of retinal at different concentrations. The spectre does not look reliable]] | + | [[Image: Team-SDU-denmark-Beta-carotene standarts.png|thumb|350px|left|'''Figure 2:''' UV-VIS of beta-carotene at different concentrations. The spectre looks reliable]][[Image: Team-SDU-denmark-Retinal standarts.png|thumb|315px|center|'''Figure 3:'''UV-VIS of retinal at different concentrations. The spectre does not look reliable]] |
<br> | <br> | ||
The spectrum for the Beta-carotene looks normal and reliable, the standards for retinal however doesn’t look reliable. The spectra for the three lowest concentrations appears to be distorted and implies that something is interfering with the measurement.<br> | The spectrum for the Beta-carotene looks normal and reliable, the standards for retinal however doesn’t look reliable. The spectra for the three lowest concentrations appears to be distorted and implies that something is interfering with the measurement.<br> | ||
| Line 44: | Line 44: | ||
MG1655 containing PSB1A2 with K274210 or PSB1C3 with K343005 were both under a constitutively active promotor<br> | MG1655 containing PSB1A2 with K274210 or PSB1C3 with K343005 were both under a constitutively active promotor<br> | ||
The measurements were preformed on cells after 20 hours of growth. The resulting graphs is presented beneath the text.<br> | The measurements were preformed on cells after 20 hours of growth. The resulting graphs is presented beneath the text.<br> | ||
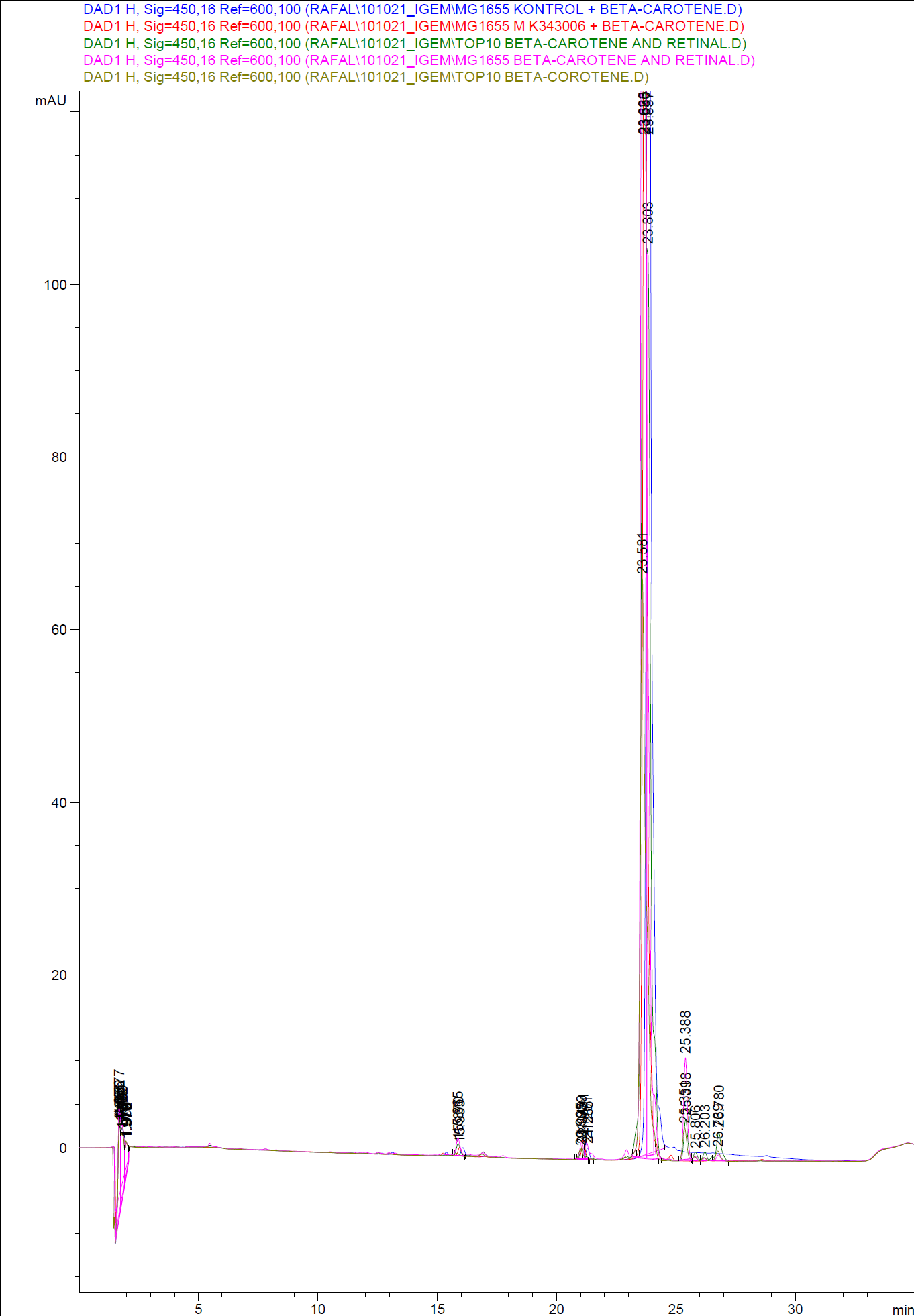
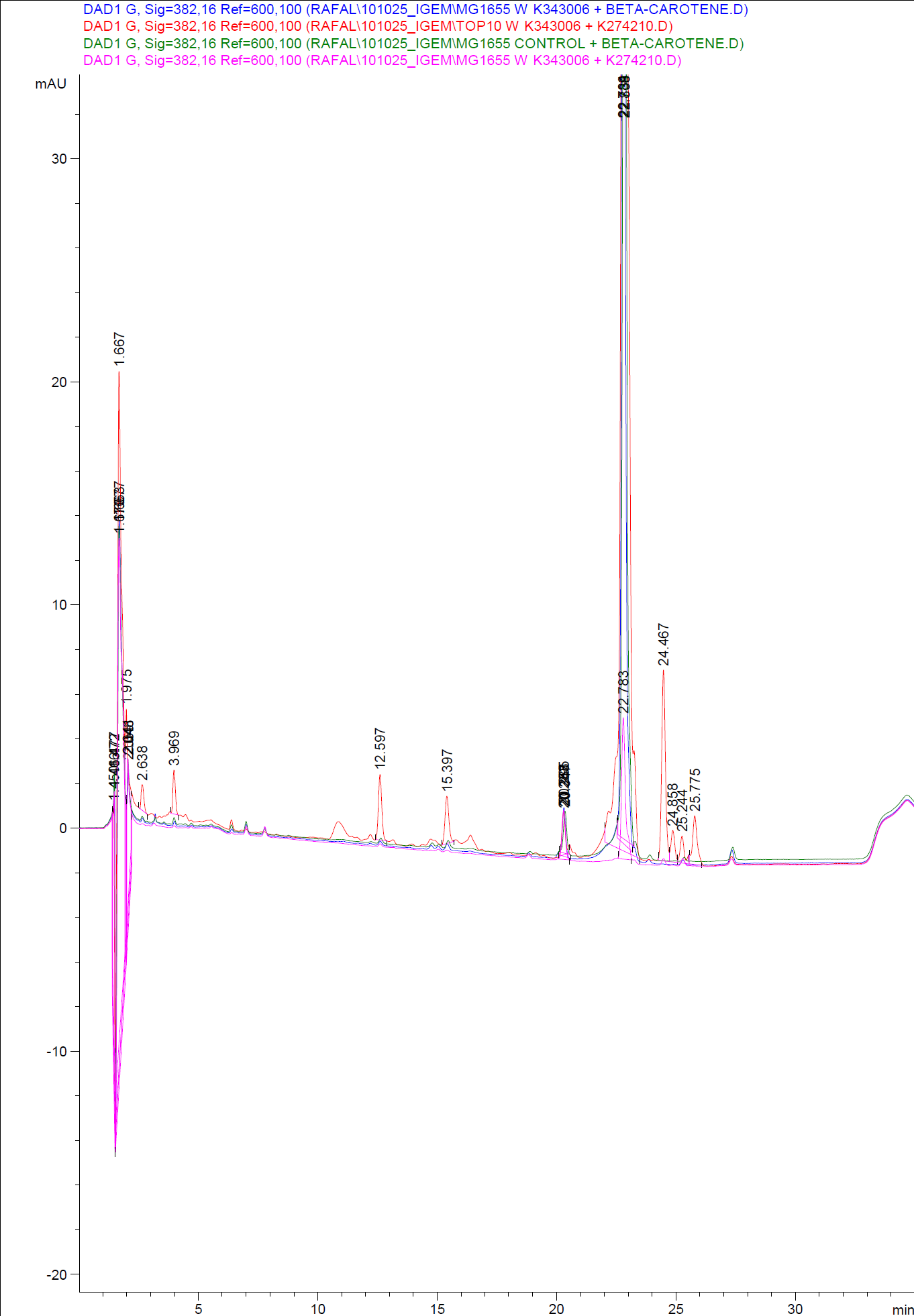
| - | [[Image:Team SDU-Denmark-HPLC samples2.png|thumb|center|300px|HPLC analysis of samples from the 19th oct. 2010 Beta-carotene Peaks are clearly present, but no retinal peaks are observed. Measurements were made after 20 hours of growth]] | + | [[Image:Team SDU-Denmark-HPLC samples2.png|thumb|center|300px|'''Figure 4:''' HPLC analysis of samples from the 19th oct. 2010 Beta-carotene Peaks are clearly present, but no retinal peaks are observed. Measurements were made after 20 hours of growth]] |
| - | [[Image:Team SDU-Denmark-HPLC samples1.png|thumb|center|300px|HPLC analysis of samples from the 22nd oct. 2010 Beta-carotene Peaks are clearly pressent, but no retinal peaks are observed. Measurements were made after 20 hours of growth]]<br> | + | [[Image:Team SDU-Denmark-HPLC samples1.png|thumb|center|300px|'''Figure 5:''' HPLC analysis of samples from the 22nd oct. 2010 Beta-carotene Peaks are clearly pressent, but no retinal peaks are observed. Measurements were made after 20 hours of growth]]<br> |
When looking at the retinal and beta-carotene standards analysed on the HPLC, peaks for both chemicals are clearly present. The retinal retention time is 3,480-3,490 minutes and the beta-carotene retention time is 23,300-23,600 minutes.<br> | When looking at the retinal and beta-carotene standards analysed on the HPLC, peaks for both chemicals are clearly present. The retinal retention time is 3,480-3,490 minutes and the beta-carotene retention time is 23,300-23,600 minutes.<br> | ||
The spectra for the first run of samples show large amounts of beta-carotene, smaller amounts of which also are products produced by the K274210 biobrick or absorbed form the media in the cells where the K274210 biobrick is absent.<br> | The spectra for the first run of samples show large amounts of beta-carotene, smaller amounts of which also are products produced by the K274210 biobrick or absorbed form the media in the cells where the K274210 biobrick is absent.<br> | ||
| Line 54: | Line 54: | ||
<br> | <br> | ||
In order to assess and quantify the results a standard dilution of beta-carotene and retinal was made and measured: 10 µM, 5 µM, 1 µM, 2 µM, 0,5 µM. The standard dilutions were also measured on the HPLC with the same injection volume and program as the samples. The resulting graphs are presented beneath <br> | In order to assess and quantify the results a standard dilution of beta-carotene and retinal was made and measured: 10 µM, 5 µM, 1 µM, 2 µM, 0,5 µM. The standard dilutions were also measured on the HPLC with the same injection volume and program as the samples. The resulting graphs are presented beneath <br> | ||
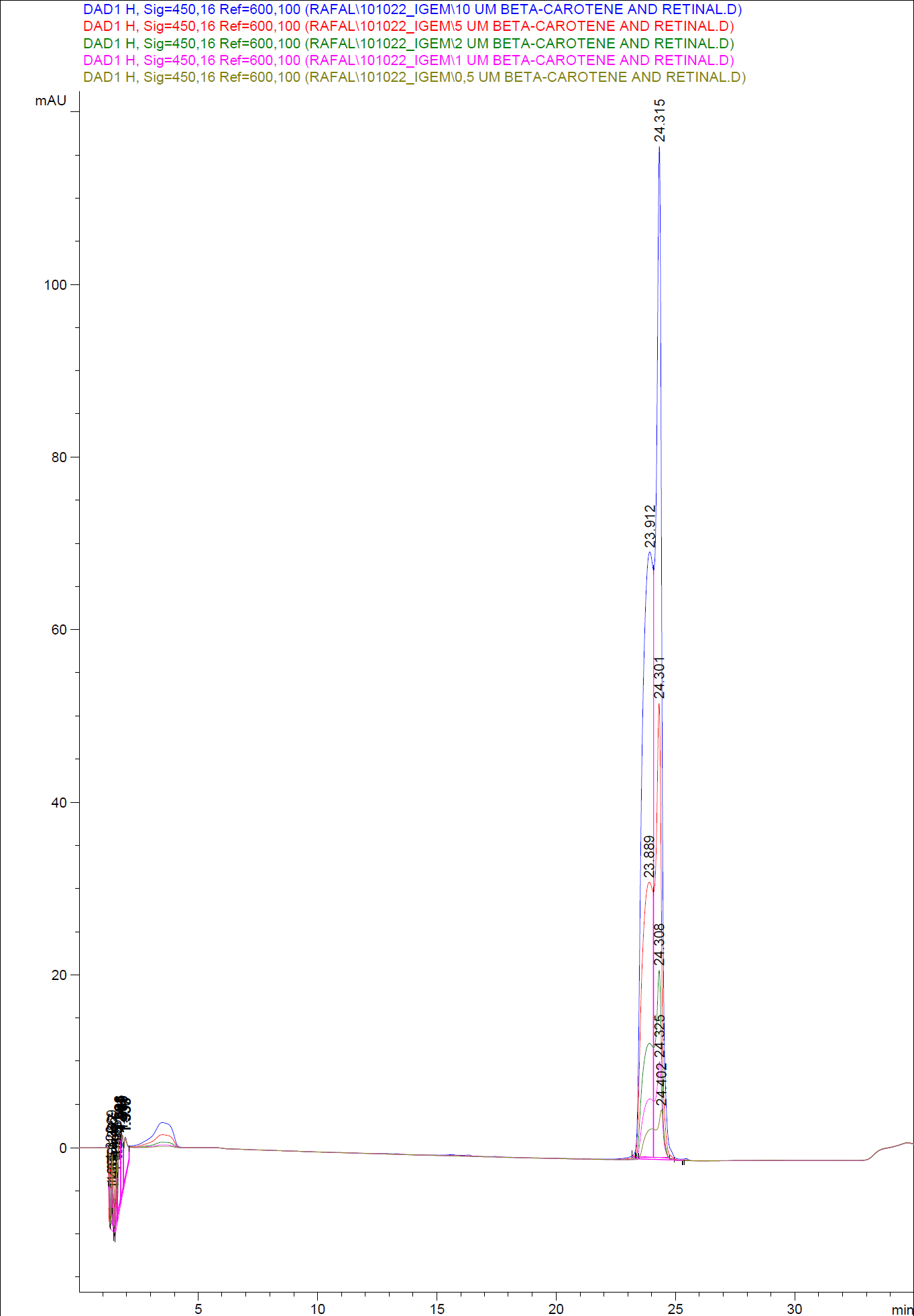
| - | [[Image:Team SDU-Denmark-HPLC standards1.png|400px|thumb|center| HPLC of standard dilutions of beta-carotene and retinal]] | + | [[Image:Team SDU-Denmark-HPLC standards1.png|400px|thumb|center| '''Figure 6:''' HPLC of standard dilutions of beta-carotene and retinal]] |
<br> | <br> | ||
When doing an HPLC analysis it is possible to quantify the amounts of the chemical compounds in the samples.<br> | When doing an HPLC analysis it is possible to quantify the amounts of the chemical compounds in the samples.<br> | ||
| Line 61: | Line 61: | ||
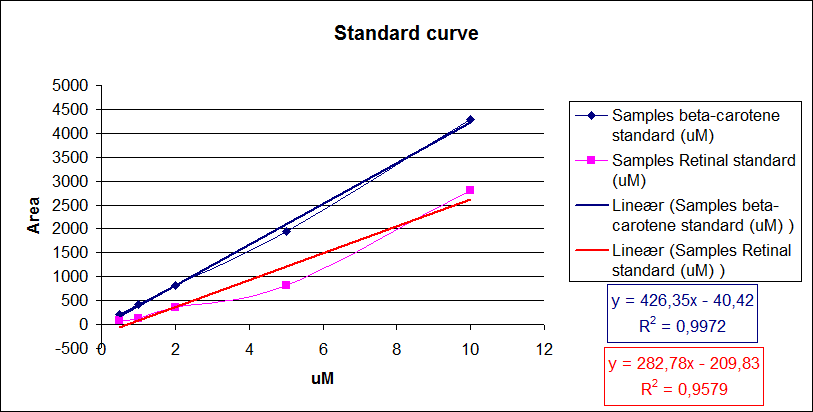
Now it is possible to calculate unknown concentrations from samples using the regression line from the standard curves.<br> | Now it is possible to calculate unknown concentrations from samples using the regression line from the standard curves.<br> | ||
The results for our HPLC analysis can be seen beneath the text.<br> | The results for our HPLC analysis can be seen beneath the text.<br> | ||
| - | [[Image:Team-SDU-Denmark-Amount of beta-carotene and retinal i standards.png|thumb|400px|center|Standard curve of area under the HPLC peaks against beta-carotene and retinal concentration. Regression lines were made.]] | + | [[Image:Team-SDU-Denmark-Amount of beta-carotene and retinal i standards.png|thumb|400px|center|'''Figure 7:''' Standard curve of area under the HPLC peaks against beta-carotene and retinal concentration. Regression lines were made.]] |
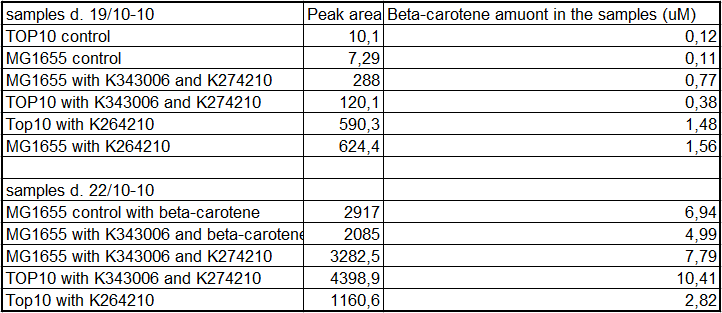
From the equations for the regression line it is possible to calculate the amount of beta-carotene produced by bacteria grown for 20 hours in LB media.<br> | From the equations for the regression line it is possible to calculate the amount of beta-carotene produced by bacteria grown for 20 hours in LB media.<br> | ||
| - | [[Image:Team-SDU-Denmark-Beta-carotene amounts.png|400px|thumb|center|Tabel | + | [[Image:Team-SDU-Denmark-Beta-carotene amounts.png|400px|thumb|center|'''Tabel 1:''' Showing calculatet beta-carotene content in samples]] |
<br> | <br> | ||
All data can be seen under [https://2010.igem.org/Image:Team-SDU-denmark-HPLC_determination_af_beta-carotene_and_retinal_production.zip Raw data]<br> | All data can be seen under [https://2010.igem.org/Image:Team-SDU-denmark-HPLC_determination_af_beta-carotene_and_retinal_production.zip Raw data]<br> | ||
| Line 69: | Line 69: | ||
=== Stability assay === | === Stability assay === | ||
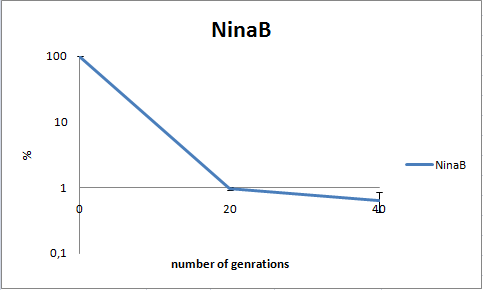
To determine the stability of our pSB1C3-K343006 plasmid, a stability experiment was carried out according to protocol [[https://2010.igem.org/Team:SDU-Denmark/protocols#Stability_assay SA1.1]]. ''E.coli'' MG1655/pSB1C3-K343006 was grown in LB media without chloramphenicol, whereby no selection pressure is exerted on the bacteria. Dilutions of the culture was spreaded onto LA plates and LA plates with 35µg/mL chloramphenicol, respectively, and the colony forming units (CFU) was determined for each plate. The CFU for the LA plates represents the total amount of bacteria in the culture, and the CFU of LA plates with chloramphenicol corresponds to the amount of plasmid carrying bacteria. The percentage of the total amount of bacteria carrying the plasmid was plotted in a semi-logarithmic graph as a function of number of generations.<br><br> | To determine the stability of our pSB1C3-K343006 plasmid, a stability experiment was carried out according to protocol [[https://2010.igem.org/Team:SDU-Denmark/protocols#Stability_assay SA1.1]]. ''E.coli'' MG1655/pSB1C3-K343006 was grown in LB media without chloramphenicol, whereby no selection pressure is exerted on the bacteria. Dilutions of the culture was spreaded onto LA plates and LA plates with 35µg/mL chloramphenicol, respectively, and the colony forming units (CFU) was determined for each plate. The CFU for the LA plates represents the total amount of bacteria in the culture, and the CFU of LA plates with chloramphenicol corresponds to the amount of plasmid carrying bacteria. The percentage of the total amount of bacteria carrying the plasmid was plotted in a semi-logarithmic graph as a function of number of generations.<br><br> | ||
| - | [[Image:Team-SDU- NinaB stab2.png|400px|thumb|center|Stability assay of ''E. coli'' strain MG1655-pSB1C3-K343006 showing that almost all cells have shed their plasmids within 20 generations, however some cells kept the plasmids for up to 40 generations.All data can be seen under [https://static.igem.org/mediawiki/2010/3/39/Team-SDU-_NinaB_stability_assay.ZIP Raw data]]]<br><br> | + | [[Image:Team-SDU- NinaB stab2.png|400px|thumb|center|'''Figure 8:''' Stability assay of ''E. coli'' strain MG1655-pSB1C3-K343006 showing that almost all cells have shed their plasmids within 20 generations, however some cells kept the plasmids for up to 40 generations.All data can be seen under [https://static.igem.org/mediawiki/2010/3/39/Team-SDU-_NinaB_stability_assay.ZIP Raw data]]]<br><br> |
As seen in the graph, almost all of the bacteria had lost the plasmid after 20 generations, suggesting that the plasmid is only stable within the cell for a few generations (<20). This is presumably due to the strain brought upon the bacteria by the plasmid. Thereby when the bacteria are carrying a high-copy plasmid like pSB1C3-K343006 it is plausible that the bacteria will quickly lose the plasmid when no longer exposed to a selection pressure. | As seen in the graph, almost all of the bacteria had lost the plasmid after 20 generations, suggesting that the plasmid is only stable within the cell for a few generations (<20). This is presumably due to the strain brought upon the bacteria by the plasmid. Thereby when the bacteria are carrying a high-copy plasmid like pSB1C3-K343006 it is plausible that the bacteria will quickly lose the plasmid when no longer exposed to a selection pressure. | ||
| Line 76: | Line 76: | ||
The purpose of this assay is to see if our transformants deviate from the wild type in growth rate. In the growth measurement assay we have measured OD at 550 nm every hour for 12 hours and at hour 24. In the experimental setup we used, no lag phase was observed in any of the measurements. | The purpose of this assay is to see if our transformants deviate from the wild type in growth rate. In the growth measurement assay we have measured OD at 550 nm every hour for 12 hours and at hour 24. In the experimental setup we used, no lag phase was observed in any of the measurements. | ||
The graph below shows the growth of our wild type E. coli strain MG1655 and the MG1655/pSB1C3-K343006, respectively.<br><br> | The graph below shows the growth of our wild type E. coli strain MG1655 and the MG1655/pSB1C3-K343006, respectively.<br><br> | ||
| - | [[Image: Team SDU-Denmark OD WT+NinaB.JPG|400px | + | [[Image: Team SDU-Denmark OD WT+NinaB.JPG|400px|thumb|center|'''Figure 9:''' Growth curve showing measured OD at 550nm. Samples were taken every hour over a 12 hour perion + one sample after 24 hours. In graph ''E. coli'' strain MG1655-pSB1C3-k343006 is compaired with Wild type ''E. coli'' strain MG1655 showing no significant difference between the three graphs. No Lag phase is seen.All data can be seen under [https://static.igem.org/mediawiki/2010/6/68/Team_SDU-Denmark_Growth_rate_assay_no._2_NinaB.zip Raw data]]] <br><br> |
From our data we see no significant difference between the plasmid carrying bacteria and the wild type. This can be said to be quite contradictory to our results obtained from the stability assay. The transitory stability of pSB1C3-K343006 suggests that it is highly unfavorable for the bacteria, wherefore it might be expected that the growth of the bacteria containing this plasmid would be affected. Thus, however large a disadvantage the plasmid pose to the bacteria, their growth are not significantly influenced by the plasmid. The added reproduction load due to the plasmids, might also prolong the lag phase of the bacteria. Whether this is the case can not be concluded based on this experiment as no lag phase was observed in this experiment. | From our data we see no significant difference between the plasmid carrying bacteria and the wild type. This can be said to be quite contradictory to our results obtained from the stability assay. The transitory stability of pSB1C3-K343006 suggests that it is highly unfavorable for the bacteria, wherefore it might be expected that the growth of the bacteria containing this plasmid would be affected. Thus, however large a disadvantage the plasmid pose to the bacteria, their growth are not significantly influenced by the plasmid. The added reproduction load due to the plasmids, might also prolong the lag phase of the bacteria. Whether this is the case can not be concluded based on this experiment as no lag phase was observed in this experiment. | ||
Latest revision as of 01:46, 28 October 2010
 "
"