Team:SDU-Denmark/K343004
From 2010.igem.org
(Difference between revisions)
(→Scanning Electron microscopy) |
|||
| (One intermediate revision not shown) | |||
| Line 5: | Line 5: | ||
== K343004 == | == K343004 == | ||
The FlhDC operon is the master regulator of flagella synthesis. A more detailed description of the operon can be found [https://2010.igem.org/Team:SDU-Denmark/project-t#Hyperflagellation here] <br> | The FlhDC operon is the master regulator of flagella synthesis. A more detailed description of the operon can be found [https://2010.igem.org/Team:SDU-Denmark/project-t#Hyperflagellation here] <br> | ||
| - | In our system the purpose of the composite part is to | + | In our system the purpose of the composite part is to hyperflagellate our cells so that a grater force can be generated in the microtubes. The FlhDC operon is naturally found in the genome of ''E. coli'' strain MG1655. We extracted the operon and introduced a silent mutation (T to C) at position 822 in the coding sequence because without the mutation this site is a Pst1 digestion site and it would therefore constitute problems when assembling the composite part. <br> We have made three FlhDC parts:<br> [http://partsregistry.org/Part:BBa_K343100 K343100] is the coding sequence of the native FlhDC operon containing the Pst1 digestion site <br> [http://partsregistry.org/Part:BBa_K343000 K343000] is the coding sequence of the mutated FlhDC operon <br> [http://partsregistry.org/Part:BBa_K343004 K343004] is the composite part containing the TetR repressed promoter (constitutive when no TetR is present) + RBS (J13002), the K343000 part and the double terminator (B0015). <br> |
| - | The composite part is | + | The composite part is characterized firstly by using a motility assay and secondly by measuring plasmid stability and cell growth. <br><br> |
===Motility assay=== | ===Motility assay=== | ||
| - | The purpose of this experiment is to test the motility of the transformed cells containing either pSB1C3-K343004 or pSB3K3-K343004. We also want to test if it makes a difference in the motility whether the bacteria contain low-medium | + | The purpose of this experiment is to test the motility of the transformed cells containing either pSB1C3-K343004 or pSB3K3-K343004. We also want to test if it makes a difference in the motility whether the bacteria contain low- medium or high-copy plasmids. <br><br> |
| - | For the motility assays we added | + | For the motility assays we added 5µl of an overnight culture to petridishes containing motility agar (LB media with 0.3% agar) instead of regular LA (Luria agar). This semi-solid media lets the bacteria swim more easily. <br> |
| - | In the assays three control plates were made: A negative control containing ''E. coli'' strain '''DH5alpha''' which does not express flagella and therefore movement in the media should be minimal. A positive control containing ''E. coli'' strain H10407 which is a hyper flagellated class II pathogen, these bacteria should show high motility. | + | In the assays three control plates were made: A negative control containing ''E. coli'' strain '''DH5alpha''' which does not express flagella and therefore movement in the media should be minimal. A positive control containing ''E. coli'' strain H10407 which is a hyper flagellated class II pathogen, these bacteria should show high motility. Lastly a wild type ''E. coli'' strain '''MG1655''' that has about 4 flagella per cell, these cells would be expected to move farther than the DH5alpha but not as far as the positive control. <br><br> |
The plates were incubated at 37 degrees celcius for up to 48 hours. <br> | The plates were incubated at 37 degrees celcius for up to 48 hours. <br> | ||
| - | The assay was carried out three times. All three times pictures were taken after 24 hours and in two of the assays pictures were also taken after 48 hours. In two of the assays 8.5 cm petridishes was used, the purpose of these assays was to | + | The assay was carried out three times. All three times pictures were taken after 24 hours and in two of the assays pictures were also taken after 48 hours. In two of the assays 8.5 cm petridishes was used, the purpose of these assays was to see if the motility of our transformants differed from the control strains. In the third assay we used 13.5 cm petridishes to observe the transformants swimming ability compared to the wild type ''E. coli'' strain MG1655 since the motility of the transformed cells seem to surpass the size of the 8.5 cm petri dishes. <br><br> |
| - | All the pictures taken after 24 hours are shown in the last part of this section while the pictures taken after 48 hours are shown and described. <br> | + | All the pictures taken after 24 hours are shown in the last part of this section while the pictures taken after 48 hours are shown and described here. <br> |
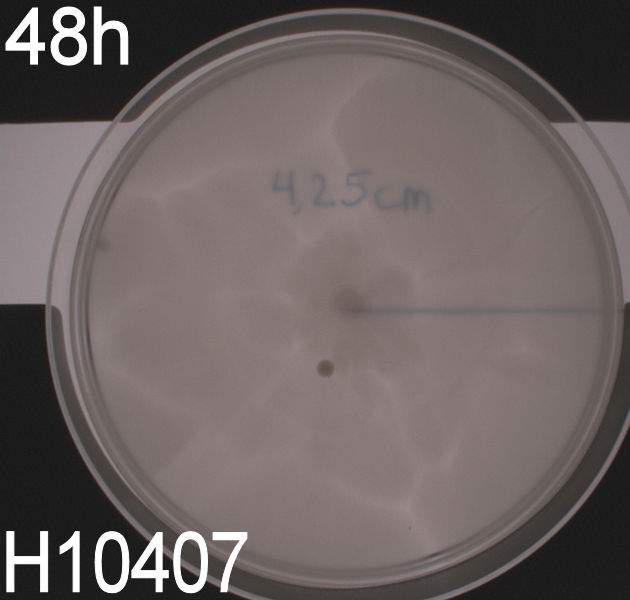
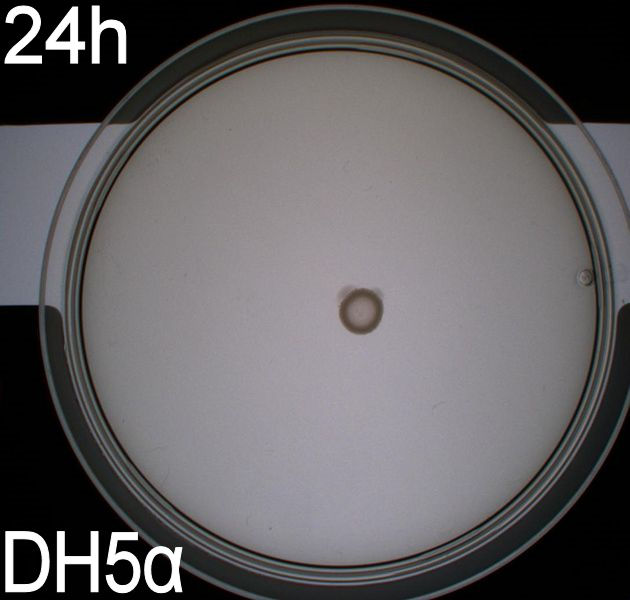
''' WT, negative- and positive-control after 48 hours:''' <br> | ''' WT, negative- and positive-control after 48 hours:''' <br> | ||
| - | [[Image:Team SDU-Denmark motility exp 3 DH5alpha.JPG|150px | + | [[Image:Team SDU-Denmark motility exp 3 DH5alpha.JPG|150px]] |
| - | [[Image:Team SDU-Denmark motility exp 3 MG1655.JPG|150px | + | [[Image:Team SDU-Denmark motility exp 3 MG1655.JPG|150px]] |
| - | [[Image:Team SDU-Denmark motility exp 3 H10407.JPG|150px | + | [[Image:Team SDU-Denmark motility exp 3 H10407.JPG|150px]] <br><br> |
| - | [[Image:Team SDU-Denmark motility exp 3 pSB1C3-K343004.JPG|300px|thumb|left|Motility of ''E. coli'' strain MG1655-pSB1C3-K343004 after 48 hours. Cells were grown in a 37 degrees Celcius incubator.]] | + | [[Image:Team SDU-Denmark motility exp 3 pSB1C3-K343004.JPG|300px|thumb|left|'''Figure 1:''' Motility of ''E. coli'' strain MG1655-pSB1C3-K343004 after 48 hours. Cells were grown in a 37 degrees Celcius incubator.]] |
| - | [[Image:Team SDU-Denmark motility exp 3 pSB3K3-K343004.JPG|300px|thumb|center|Motility of ''E. coli'' strain MG1655-pSB3K3-K343004 after 48 hours. Cells were grown in a 37 degrees Celcius incubator.]]<br><br> | + | [[Image:Team SDU-Denmark motility exp 3 pSB3K3-K343004.JPG|300px|thumb|center|'''Figure 2:''' Motility of ''E. coli'' strain MG1655-pSB3K3-K343004 after 48 hours. Cells were grown in a 37 degrees Celcius incubator.]]<br><br> |
| - | In all three assays we saw that the control plates containing no antibiotics were contaminated with other bacterial colonies. We also saw that the negative control has low motility but they are not immotile. After 24 hours the negative control has not moved much but after 48 hours the length from center to edge of the colony is 1.5cm. The wild type has moved a bit farther than the negative control after 24 hours and after 48 hours the difference from center to edge of the wild type is 2.5cm. The positive control moved about as far in 24 hours as the wild type did in 48 which and after 48 hours it had moved to the edges of the petridish (4.25cm). | + | In all three assays we saw that the control plates containing no antibiotics were contaminated with other bacterial colonies. We also saw that the negative control has low motility but they are not immotile. After 24 hours the negative control has not moved much but after 48 hours the length from center to edge of the colony is 1.5cm. The wild type has moved a bit farther than the negative control after 24 hours and after 48 hours the difference from center to edge of the wild type is 2.5cm. The positive control moved about as far in 24 hours as the wild type did in 48 which and after 48 hours it had moved to the edges of the petridish (4.25cm). This was as expected because of their hyper flagellation. <br><br> |
| - | The ''E. Coli'' strain MG1655 | + | The ''E. Coli'' strain MG1655 transformed with pSB1C3-K343004 shows that after 24 hours these bacteria have moved farther than the wild type and the negative control. The ''E. coli'' strain MG1655/pSB3K3-K343004 has moved the farthest of all 5 cultures after 24 hours. This motility pattern is observed in all three assays. The cells containing the pSB3K3 plasmid shows a uniform circle whereas the cells transformed with the pSB1C3 plasmid shows a budding-pattern spreading from the center. After 48 hours the pSB1C3 cells are still not covering the entire plate. <br><br> |
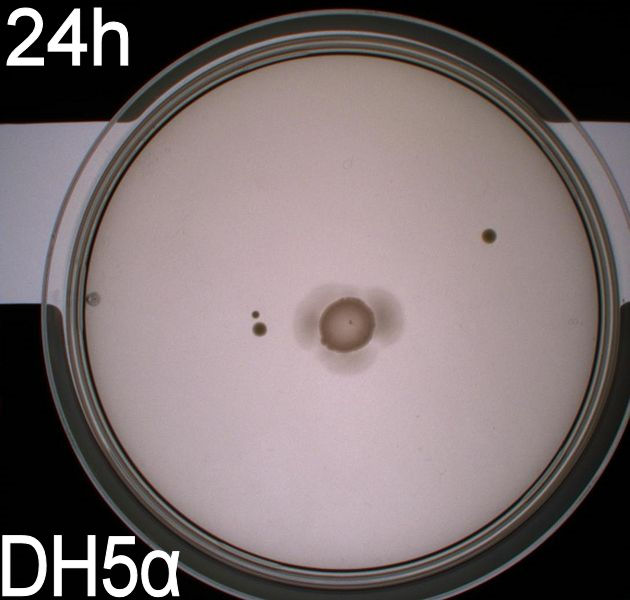
'''''Pictures of plates after 24 hours''''' <br> | '''''Pictures of plates after 24 hours''''' <br> | ||
'''Experiment 1.''' <br> | '''Experiment 1.''' <br> | ||
| Line 38: | Line 38: | ||
''' Conclusion''' <br> | ''' Conclusion''' <br> | ||
From the pictures above we can see that the bacteria containing our part is much more motile than the wild type and the negative control. We assume this is caused by overexpression of the FlhDC master flagella operon which leads to hyperflagellation of the cells. <br> | From the pictures above we can see that the bacteria containing our part is much more motile than the wild type and the negative control. We assume this is caused by overexpression of the FlhDC master flagella operon which leads to hyperflagellation of the cells. <br> | ||
| - | The pictures taken after 24 hours show that bacteria with pSB1C3-K343004 have not moved as far as the bacteria containing pSB3K3-K343004. pSB1C3 is a high copy plasmid while pSB3K3 is a low-medium copy plasmid. The promoter in K343004 is a constitutive promoter (tetR | + | The pictures taken after 24 hours show that bacteria with pSB1C3-K343004 have not moved as far as the bacteria containing pSB3K3-K343004. pSB1C3 is a high copy plasmid while pSB3K3 is a low-medium copy plasmid. The promoter in K343004 is a constitutive promoter (tetR repressed promoter). Bacteria containing a high copy plasmid with a constitutive promoter are more metabolically challenged than bacteria containing a low- or medium-copy plasmid with a constitutive promoter because of the higher number of plasmids per cell. Therefore the high copy plasmid bacteria move slower than low- or medium-copy plasmid bacteria. However, after 48 hours there was only a small different in the bacterial migration. <br><br> |
| - | + | === Phase contrast of overnight cultures=== | |
| - | === Phase contrast of | + | |
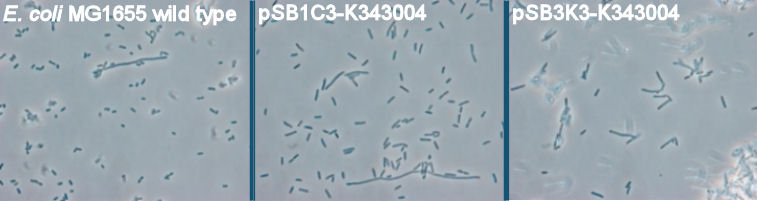
The purpose of the phase contrast microscopy is to see if the transformants are visibly different from the wild type. Since the FlhDC operon is not only an important part of the flagella synthesis but is also coupled to cell metabolism [[https://2010.igem.org/Team:SDU-Denmark/project-p#References 3]], it might be possible to see difference in morphology caused by an alterations in the cell cycle G2-phase. <br><br> | The purpose of the phase contrast microscopy is to see if the transformants are visibly different from the wild type. Since the FlhDC operon is not only an important part of the flagella synthesis but is also coupled to cell metabolism [[https://2010.igem.org/Team:SDU-Denmark/project-p#References 3]], it might be possible to see difference in morphology caused by an alterations in the cell cycle G2-phase. <br><br> | ||
| - | + | [[Image:Team-SDU-Denmark-Flagella-PCM-1.jpg|630px|thumb|center|'''Figure 3:''' Phase contrast pictures of wild type MG1655, MG1655/pSB1C3-K343004 and MG1655/pSB3K3-K343004. The bacterias was incubated ON at 37 degress and LB-media]] <br><br> | |
| - | [[Image:Team-SDU-Denmark-Flagella-PCM-1.jpg|630px|thumb|center|Phase contrast pictures of wild type MG1655, MG1655/pSB1C3-K343004 and MG1655/pSB3K3-K343004. The bacterias was incubated ON at 37 degress and LB-media]] <br><br> | + | |
The pictures clearly show a difference in morphology of the wild type (picture 1) and the two transformants (picture 2-pSB1C3 and 3-pSB3K3). The wild type cells are small cells, while the transformants are obviously larger, which indicates a lower metabolism of the transformants. Cells with impaired metabolism divide slower and are larger than cells with normal metabolism (wild type), since it takes the impaired cells longer to reach the mitotic phase of the cell cycle. <br> <br> | The pictures clearly show a difference in morphology of the wild type (picture 1) and the two transformants (picture 2-pSB1C3 and 3-pSB3K3). The wild type cells are small cells, while the transformants are obviously larger, which indicates a lower metabolism of the transformants. Cells with impaired metabolism divide slower and are larger than cells with normal metabolism (wild type), since it takes the impaired cells longer to reach the mitotic phase of the cell cycle. <br> <br> | ||
===Flagella staining=== | ===Flagella staining=== | ||
<br> | <br> | ||
| - | When the ''E.coli'' strain MG1655 | + | When the ''E. coli'' strain MG1655 is transformed with pSB1C3-K343004 an increased flagella production is expected. The idea was to get a quantitative measurement of the increased flagella production by comparing the observed number flagella on the wild type MG1655 and the transformant. We tried to stain the flagella using silver staining and afterwards examined the bacteria under the microscope. The staining procedure was started before the composite part, K343004, was finished so that we could optimize the staining protocol and become really good at staining the flagella. We started with staining of DH5α, an ''E. coli'' strain that does not express flagella (negative control) and H10407, a hyperflagellated ''E. coli'' (positive control). The bacteria were grown on agar plates overnight and stained with this protocol [https://2010.igem.org/Team:SDU-Denmark/protocols#FS1.2 FS1.2]. <br> <br> |
Unfortunately it was not possible to see a clear and significant difference between the positive and negative control. Though at some places it could look like flagella on the positive control but it was never enough to determine a difference. After repeating the protocol four times, each time with multiple samples, we decided to reject this method, and another approach for characterizing our biobrick was employed. <br> | Unfortunately it was not possible to see a clear and significant difference between the positive and negative control. Though at some places it could look like flagella on the positive control but it was never enough to determine a difference. After repeating the protocol four times, each time with multiple samples, we decided to reject this method, and another approach for characterizing our biobrick was employed. <br> | ||
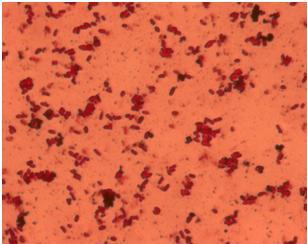
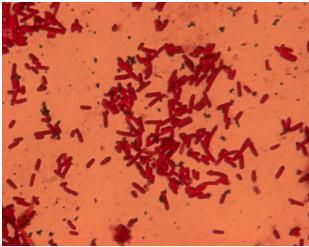
| - | + | [[Image:Team_SDU-Denmark_Silver_staining_of_H10407_a_positive_control..JPG|300px|thumb|left|'''Figure 4:''' Silver staining of ''E. coli'' strain H10407 which should be hyperflagellated. However, no .]][[Image:Team_SDU-Denmark_Silver_staining_of_DH5α_the_negative_control.JPG|300px|thumb|center|'''Figure 5:'''Silver staining of DH5alpha: a negative control]]<br> | |
| - | [[Image:Team_SDU-Denmark_Silver_staining_of_H10407_a_positive_control..JPG|300px|thumb|left|Silver staining of ''E. coli'' strain H10407 which should be hyperflagellated. However, no .]][[Image:Team_SDU-Denmark_Silver_staining_of_DH5α_the_negative_control.JPG|300px|thumb|center|Silver staining of DH5alpha: a negative control]]<br> | + | |
=== Scanning Electron microscopy === | === Scanning Electron microscopy === | ||
<br> | <br> | ||
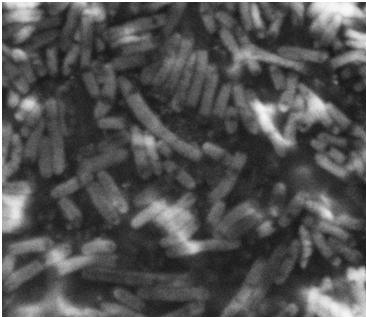
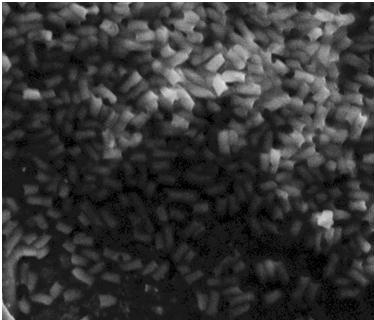
| - | After the failed | + | After the failed staining of the flagella we tried to visualize the flagella with scanning electron microscopy (SEM). The bacteria were grown overnight in liquid cultures (5 ml LB-media). The bacteria were diluted to approximately 10^6 cells in 10 µl. At that time we did not have our K343004 composite part so therefore we only tried SEM with the negative control strain DH5alpa and wild type MG1655. We did this as a preliminary work so we would be ready to do microscopy on the MG1655 cells containing the composite part when it was ready. Unfortunately the limited resolution and magnitude of the SEM at our disposal made it practically impossible to visualize any flagella in the microscope. Thus if this approach is to be used for characterization, a SEM of higher magnitude and resolution is required. Our plan was instead to use a transmission electron microscope but unfortunately it was been unfunctional for that last week of iGEM. <br> |
| - | The pictures below are SEM pictures the negative control strain DH5alpha and wild type MG1655.<br> <br> | + | The pictures below are SEM pictures of the negative control strain DH5alpha and wild type MG1655.<br> <br> |
| - | [[Image:Team SDU-Denmark_DH5alpha_MAG_5.0_kx_and_EHT_7.5_kV.JPG|thumb|left|300px|SEM picture of DH5alpa a negative control that should not express flagella]][[Image:Team_SDU-Denmark_Mg1655_MAG5kx_and_EHT_7.5kV.JPG|thumb|center|300px|SEM picture of the ''E. coli'' strain MG1655 wild type this strain of bacteria should express approximately 4 flagella per cell.However, no flagella are visable]] | + | [[Image:Team SDU-Denmark_DH5alpha_MAG_5.0_kx_and_EHT_7.5_kV.JPG|thumb|left|300px|'''Figure 6:''' SEM picture of DH5alpa a negative control that should not express flagella]][[Image:Team_SDU-Denmark_Mg1655_MAG5kx_and_EHT_7.5kV.JPG|thumb|center|300px|'''Figure 7:''' SEM picture of the ''E. coli'' strain MG1655 wild type this strain of bacteria should express approximately 4 flagella per cell. However, no flagella are visable]] <br> |
| - | + | ||
===Stability assay=== | ===Stability assay=== | ||
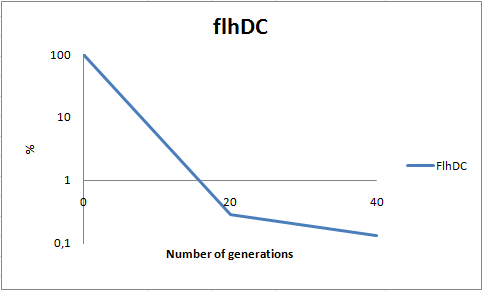
| - | To | + | To determine the stability of our pSB1C3-K343004 plasmid, a stability experiment was carried out according to protocol [[https://2010.igem.org/Team:SDU-Denmark/protocols#Stability_assay SA1.1]]. ''E. coli'' MG1655/pSB1C3-K343004 was grown in LB media without chloramphenicol, whereby no selection pressure is exerted on the bacteria. Dilutions of the culture was spread onto LA plates and LA plates with 35µg/mL chloramphenicol, respectively, and the colony forming units (CFU) was determined for each plate. The CFU for the LA plates represents the total amount of bacteria in the culture, and the CFU of LA plates with chloramphenicol corresponds to the amount of plasmid carrying bacteria. The percentage of the total amount of bacteria carrying the plasmid was plotted in a semi-logarithmic graph as a function of number of generation.<br><br> |
| - | [[Image:Team-SDU-DC stab 2.png|thumb|400px|center| Stability assay of ''E. coli'' strain MG1655-pSB1C3-K343004 showing that almost all cells have shed their plasmids within 20 generations, however some cells kept the plasmids for up to 40 generations. All data can be seen under [https://static.igem.org/mediawiki/2010/0/09/Team-SDU-_flhDC_stability_assay.ZIP Raw data]]] | + | [[Image:Team-SDU-DC stab 2.png|thumb|400px|center| '''Figure 8:''' Stability assay of ''E. coli'' strain MG1655-pSB1C3-K343004 showing that almost all cells have shed their plasmids within 20 generations, however some cells kept the plasmids for up to 40 generations. All data can be seen under [https://static.igem.org/mediawiki/2010/0/09/Team-SDU-_flhDC_stability_assay.ZIP Raw data]]] |
<br><br> | <br><br> | ||
| - | As seen in the graph, almost all of the bacteria had | + | As seen in the graph, almost all of the bacteria had lost the plasmid after 20 generations, suggesting that the plasmid is only stable within the cell for a few generations (<20). This is presumably due to the strain brought upon the bacteria by the plasmid. Thereby when the bacteria are carrying a high-copy plasmid like pSB1C3-K343004 it is to be expected that the bacteria will quickly lose the plasmid when no longer exposed to a selection pressure. |
| - | It is likely to believe that pSB3C5-K343004, since being a low-copy plasmid, will not | + | It is likely to believe that pSB3C5-K343004, since being a low-copy plasmid, will not exert as much strain on the bacteria, and might therefore be stable for more generations than pSB1C3-K343004. Therefore a stability assay of this plasmid might be of interest. <br> |
| - | + | ||
===Growth assay=== | ===Growth assay=== | ||
The purpose of this assay is to see if our transformants deviate from the wild type in growth rate. In the growth measurement assay we have measured OD at 550 nm every hour for 12 hours and at hour 24. In the experimental set-up we used no lag phase was observed in any of the measurements. | The purpose of this assay is to see if our transformants deviate from the wild type in growth rate. In the growth measurement assay we have measured OD at 550 nm every hour for 12 hours and at hour 24. In the experimental set-up we used no lag phase was observed in any of the measurements. | ||
| - | The graph below shows the growth of our wild type E. coli strain MG1655, the MG1655/pSB3K3-K343004 and MG1655/pSB1C3-K343004 | + | The graph below shows the growth of our wild type ''E. coli'' strain MG1655, the MG1655/pSB3K3-K343004 and MG1655/pSB1C3-K343004.<br><br> |
| - | [[Image:Team SDU-Denmark OD WT+FlhDCmutCP.JPG|400px|thumb|center|Growth curve of ''E.coli'' strain MG1655-pSB1C3-K343004 and MG1655-pSB3K3-K343004 compared with the wild type ''E. coli'' strain MG1655 showing no significant difference between the three curves. No Lag phase observed. All data can be seen under [https://static.igem.org/mediawiki/2010/c/c5/Team_SDU-Denmark_Growth_rate_assay_FlhDCmut_CP.zip Raw data]]]<br><br> | + | [[Image:Team SDU-Denmark OD WT+FlhDCmutCP.JPG|400px|thumb|center|'''Figure 9:''' Growth curve of ''E.coli'' strain MG1655-pSB1C3-K343004 and MG1655-pSB3K3-K343004 compared with the wild type ''E. coli'' strain MG1655 showing no significant difference between the three curves. No Lag phase observed. All data can be seen under [https://static.igem.org/mediawiki/2010/c/c5/Team_SDU-Denmark_Growth_rate_assay_FlhDCmut_CP.zip Raw data]]]<br><br> |
| - | From our data we see no significant difference between the plasmid carrying bacteria and the wild type. This can be said to be quite contradictory to our results obtained from the stability assay. The transitory stability of pSB1C3-K343004 suggests that it is highly | + | From our data we see no significant difference between the plasmid carrying bacteria and the wild type. This can be said to be quite contradictory to our results obtained from the stability assay. The transitory stability of pSB1C3-K343004 suggests that it is highly unfavorable for the bacteria, wherefore it might be expected that the growth of the bacteria containing this plasmid would be affected. Thus, however much disadvantages the plasmid pose to the bacteria, their growth are not significantly influenced by the plasmid. The added reproduction load due to the plasmids, might also prolong the lag phase of the bacteria. Whether this is the case can not be concluded based on this experiment as no lag phase was observed in this experiment. |
<br><br> | <br><br> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 02:33, 28 October 2010
 "
"