Team:SDU-Denmark/Bielefeld
From 2010.igem.org
(Difference between revisions)
(→Helping another iGEM Team with characterization) |
|||
| (One intermediate revision not shown) | |||
| Line 6: | Line 6: | ||
==Helping another iGEM Team with characterization== | ==Helping another iGEM Team with characterization== | ||
== 2010 [https://2010.igem.org/Team:Bielefeld-Germany Bielefeld] brick K389016 == | == 2010 [https://2010.igem.org/Team:Bielefeld-Germany Bielefeld] brick K389016 == | ||
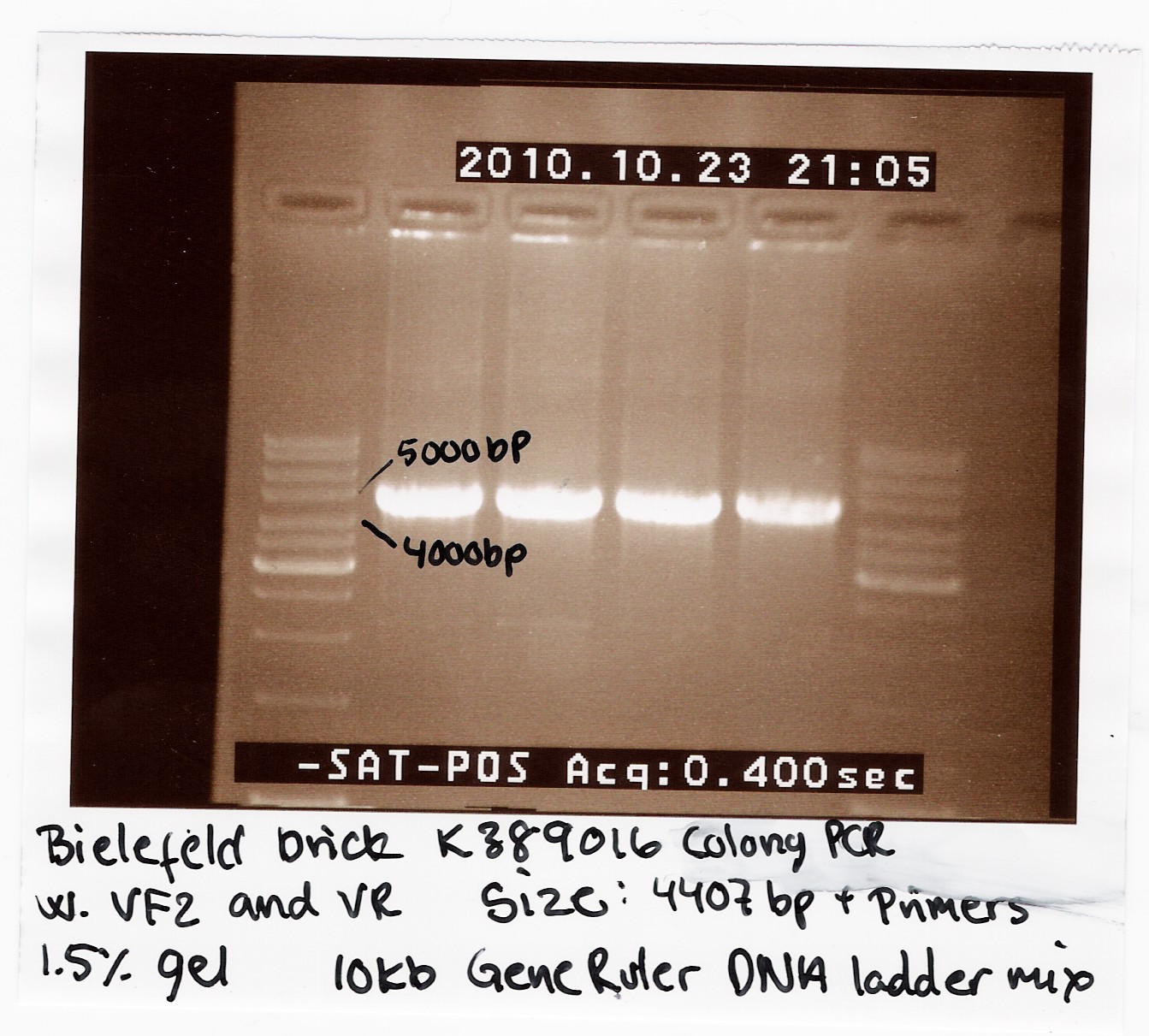
| - | [[Image:Team SDU-Denmark Bielefeld Colony PCR-1.jpg |210px|thumb|Colony-PCR of K389016]] | + | [[Image:Team SDU-Denmark Bielefeld Colony PCR-1.jpg |210px|thumb|'''Figure 1:''' Colony-PCR of K389016 using VF2, VR primers. The PCR-product was run on a 1.5% gel with 10kb GeneRuler, all bands are positioned right under 5000kb. The K389016 part has a length of 4407bp+VF2,VR]] |
| - | We helped the Bielefeld team | + | We helped the 2010 Bielefeld team with characterizing their brick K389016. It is a VirA/G receptor system with an mRFP reporter gene. Our task was to replicate their experiment which showed that acetosyringone induced the promoter and thereby increased expression of mRFP. <br><br> |
We recieved a miniprep with [http://partsregistry.org/Part:BBa_K389016 K389016] in pSB1C3 together with a list of experiments and characterization assays the Bielefeld team would like us to carry out. <br> | We recieved a miniprep with [http://partsregistry.org/Part:BBa_K389016 K389016] in pSB1C3 together with a list of experiments and characterization assays the Bielefeld team would like us to carry out. <br> | ||
We transformed the plasmids into ''E. coli'' strain MG1655 and ran a colony-PCR of four different colonies the following day. Over night cultures of the colonies was made and used for fluorescence measurements. The picture shows that cells in all four colonies contained K389016 in pSB1C3. | We transformed the plasmids into ''E. coli'' strain MG1655 and ran a colony-PCR of four different colonies the following day. Over night cultures of the colonies was made and used for fluorescence measurements. The picture shows that cells in all four colonies contained K389016 in pSB1C3. | ||
<br><br> | <br><br> | ||
=== Characterization === | === Characterization === | ||
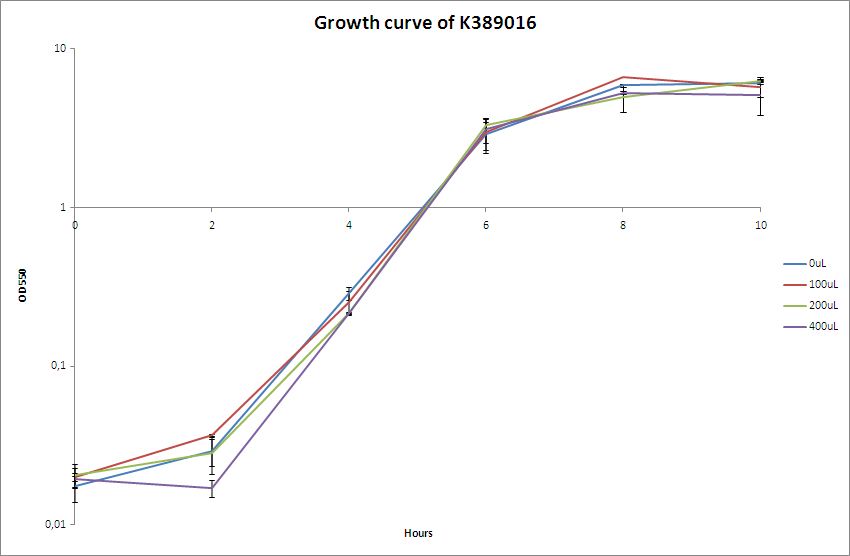
| - | [[Image:Team-SDU-Bielefeld.PNG|400px|thumb|OD550 nm of MG1655-pSB1C3-K389016]] The OD550 nm was measured every two hours for a 10 hour period. <br> | + | [[Image:Team-SDU-Bielefeld.PNG|400px|thumb|'''Figure 2:''' OD550 nm of MG1655-pSB1C3-K389016]] The OD550 nm was measured every two hours for a 10 hour period. <br> |
| - | The characterisation experiment | + | The characterisation experiment was carried out according to protocol ([https://2010.igem.org/Team:SDU-Denmark/protocols#Charactarization_of_K389016_.28VirA.2FG_reporter_device_mRFP.29 CK1.1]). The chosen transformant were grown overnight in LB-media and on the following day boosted and grown with the different acetosyringone concentrations: 0uM, 100uM, 200uM and 400uM. The culture without acetosyringone was used as a control. The cultures were incubated at 37 degrees Celsius until the cells reached stationary phase. A growth assay was carried out with measurements taken at OD<sub>550</sub> every 2 hours for 10 hours. For further knowledge see [https://static.igem.org/mediawiki/2010/d/da/Bielefeld_karakterisering_OD.zip raw data.] Parallel with the OD<sub>550</sub> measurements samples were taken and used for fluorescence measurements.<br><br> |
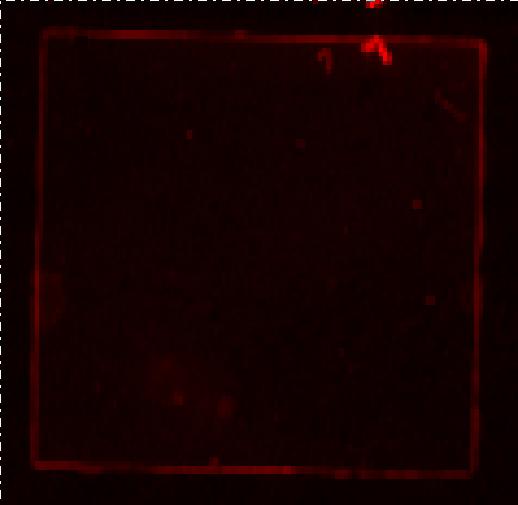
| - | [[Image:Team SDU-Denmark Bielefeld Typhoon trio.JPG|210px|thumb|left|Typhoon trio fluorescence scanning of MG1655-pSB1C3-K389016 induced with 200 uM acetosyringone. Excitation: 633nm Emittance: 670nm. No RFP detection, any red seen in the picture is | + | [[Image:Team SDU-Denmark Bielefeld Typhoon trio.JPG|210px|thumb|left|'''Figure 3:''' Typhoon trio fluorescence scanning of MG1655-pSB1C3-K389016 induced with 200 uM acetosyringone. Excitation: 633nm Emittance: 670nm. No RFP detection, any red seen in the picture is noise.]] <br> |
| - | The fluorescence measurements | + | The fluorescence measurements were carried out on a Perkin Elmer LS55 fluorescence spectrometer and the data was analysis with FL win Lab program. We did single read with excitation on 584 nm and emission 607 nm. We tried using both different excitation and emission slit size and performed the experiment twice but unfortunately we were unable to detect RFP in the cells. See the fluorescence [https://static.igem.org/mediawiki/2010/1/10/Team-sdu-denmark-Fluorescence.zip raw data] here. <br><br> |
| - | We furthermore tested the cells on a Typhoon Trio but once again we could not detect RFP. We were advised by the Bielefeld team to use excitation 584nm and | + | We furthermore tested the cells on a Typhoon Trio but once again we could not detect RFP. We were advised by the Bielefeld team to use excitation at 584nm and emission at 607nm. This was done when using the Perkin Elmer LS55 fluorescence spectrometer, but the Typhoon trio has limited settings and according to the Typhoon trio manual RFP can be detected at excitation R 633nm and emission at 670nm. <br> |
| - | The picture above shows the fluorescence scanning preformed on the Bielefeld part induced with 200uM acetosyringone. We | + | The picture above shows the fluorescence scanning preformed on the Bielefeld part induced with 200uM acetosyringone. We pelleted an over night culture of MG1655-pSB1C3-K38901 down, removed the supernatant and transferred the pellet to a glass slide and covered by a cover slide. No emissions are seen in the picture, any light seen on the picture is a result of noise. <br><br> |
| - | + | None of us have any experience in working with the fluorescence spectrometer or the part it self. Further more our fluorescence spectrometer is different than the one the Bielefeld team used, therefore the instrumental settings might be different from their’s. Because we only had two days to replicate the experiment, we did not have time to try different settings. Overall the insignificant data are probably due to our lack time, experience with the brick and not the functionality of the biobrick, K389016. | |
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 01:31, 28 October 2010
 "
"