Team:RMIT Australia/Project
From 2010.igem.org
(Prototype team page) |
(→Arginine Vasopressin) |
||
| (63 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | + | __NOTOC__ | |
| - | + | ||
<html> | <html> | ||
| - | < | + | <body style="background-color:#CCCCCC"> |
| - | + | ||
| - | + | <style> | |
| - | + | h1.firstHeading { display: none; } | |
| - | + | ||
| - | + | p {text-align: justify;} | |
| - | </ | + | |
| - | < | + | a:link { color: #FF0000; text-decoration: none} |
| - | + | a:visited { color:#FF0000; text-decoration: none} | |
| - | </ | + | a:hover { color:#FF0000; text-decoration: none} |
| + | a:active { color:#FF0000; text-decoration: none} | ||
| + | |||
| + | #bodyContent { padding: 10px auto; width: 910px; margin: auto; clear: none; } | ||
| + | |||
| + | table#team_members { text-align: justify; border: 0; } | ||
| + | table#team_members h2, table#team_members h3 { clear: both; } | ||
| + | |||
| + | |||
| + | /*-----------------------------------------------------------------------------------------------*/ | ||
| + | div.MenuBar ul li ul.DropDownMenu { | ||
| + | display: none; /* Hides all drop-down menus. */ | ||
| + | |||
| + | } | ||
| + | /* | ||
| + | li:hover works in IE7 and FF2. | ||
| + | a:hover works in IE6 and FF2. | ||
| + | a:hover breaks li:hover in FF2. | ||
| + | */ | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li ul.SideMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a ul.SideMenu { | ||
| + | display: none; /* Hides all side menus. */ | ||
| + | } | ||
| + | /*------------------------------------------------------------------------------------- Menu Bar */ | ||
| + | div.MenuBar { | ||
| + | font: arial, helvetica, sans-serif; | ||
| + | height: 30px; | ||
| + | width: 910px; | ||
| + | /*width: 100%*/ | ||
| + | margin: 0; | ||
| + | border-top: 0; | ||
| + | border-right: 0; | ||
| + | border-left: 0; | ||
| + | padding: 0; | ||
| + | background: black; | ||
| + | |||
| + | } | ||
| + | div.MenuBar ul { | ||
| + | font: arial, helvetica, sans-serif; | ||
| + | text-align: center; | ||
| + | list-style-type: none; | ||
| + | margin: 0.5em auto; | ||
| + | border: 0; | ||
| + | padding: 0; | ||
| + | background: black; | ||
| + | } | ||
| + | div.MenuBar ul li { | ||
| + | font: arial, helvetica, sans-serif; | ||
| + | display: block; | ||
| + | padding: 0; | ||
| + | margin: 0; | ||
| + | font-size: 1.3em; | ||
| + | float: left; | ||
| + | background: black; | ||
| + | text-align: center; | ||
| + | width: 107px; | ||
| + | position: relative; /* Sets the positioning context for each drop-down menu. */ | ||
| + | } | ||
| + | |||
| + | div.MenuBar ul li a { | ||
| + | font: arial, helvetica, sans-serif; | ||
| + | display: block; | ||
| + | background: black; | ||
| + | height: 22px; /* Keep height + padding-top + padding-bottom sync with the menu bar height. */ | ||
| + | color: #ffffff; | ||
| + | padding-top: 4px; | ||
| + | padding-bottom: 4px; | ||
| + | padding-left: 1em; /* Sets the left space between top-level items. */ | ||
| + | padding-right: 1em; /* Sets the right space between top-level items. */ | ||
| + | text-decoration: none; | ||
| + | } | ||
| + | |||
| + | /*------------------------------------------------------------------------------ Drop-Down Menus */ | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu { | ||
| + | display: block; | ||
| + | width: 10em; /* Drop-down menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | height: 1em; | ||
| + | padding: 1px; /* Sets the drop-down menu "effective border" width. */ | ||
| + | position: absolute; | ||
| + | top: 23px; /* Places the drop-down menu under the menu bar. | ||
| + | Keep it sync with the menu bar height. */ | ||
| + | left: 0; /* Aligns the drop-down menu to its top-level item. */ | ||
| + | background-color: black; /* Selected item. */ | ||
| + | color: #FFFFFF; | ||
| + | |||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a { | ||
| + | width: 10em; /* Keep it sync with the drop-down menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | height: 1em; | ||
| + | padding-left: 0; | ||
| + | padding-right: 0; | ||
| + | background-color: black; /* Selected item. */ | ||
| + | color: #FFFFFF; | ||
| + | } | ||
| + | ul.DropDownMenu li a span { | ||
| + | display: block; | ||
| + | padding-left: 0.75em; /* Sets the left space of each drop-down menu item. */ | ||
| + | padding-right: 0.25em; /* Sets the right space of each drop-down menu item. */ | ||
| + | text-align: right; /* Aligns the >> symbol to the right. */ | ||
| + | } | ||
| + | ul.DropDownMenu li a span span { | ||
| + | float: left; /* Aligns the text (back) to the left. */ | ||
| + | font: 12px arial, helvetica, sans-serif; /* Required for IE55. */ | ||
| + | height: 20px; | ||
| + | color: #FFFFFF; | ||
| + | } | ||
| + | /*----------------------------------------------------------------------------------- Side Menus */ | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu { | ||
| + | display: block; | ||
| + | width: 11em; /* Side menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | padding: 1px; /* Sets the side menu "effective border" width. */ | ||
| + | position: absolute; | ||
| + | top: -1px; /* Aligns the side menu to its drop-down menu item. | ||
| + | Keep it sync with the side menu "effective border" width. */ | ||
| + | left: 13em; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu li a { | ||
| + | width: 11em; /* Keep it sync with the side menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | font: 12px arial, helvetica, sans-serif; /* Required for IE55. */ | ||
| + | left: 13em; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | } | ||
| + | div.MenuBar ul li ul.DropDownMenu li ul.SideMenu li a span { | ||
| + | padding-left: 1.5em; /* Sets the left space of each side menu item. */ | ||
| + | padding-right: 0.5em; /* Sets the right space of each side menu item. */ | ||
| + | text-align: left; | ||
| + | font: 12px arial, helvetica, sans-serif; /* Required for IE55. */ | ||
| + | left: 13em; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. | ||
| + | Use MenuTailor.css to customize. */ | ||
| + | } | ||
| + | /*----------------------------------------------------------------------------- Browser Specific */ | ||
| + | * html div.MenuBar ul li a { | ||
| + | float: left; /* Required for IE55 and IE6. | ||
| + | Breaks O9. | ||
| + | Hidden (* html) from non-IE browsers. */ | ||
| + | } | ||
| + | * html ul.DropDownMenu li a:hover { | ||
| + | cursor: hand; /* Required for IE55. | ||
| + | Hidden (* html) from non-IE browsers. */ | ||
| + | } | ||
| + | ul.DropDownMenu li a:hover { | ||
| + | cursor: pointer; /* Required for IE6 and IE7. | ||
| + | Hidding it (* html) from non-IE browsers breaks IE7! | ||
| + | } | ||
| + | * html div.MenuBar a:hover { | ||
| + | text-decoration: none; /* Required for IE55 and IE6. | ||
| + | Hidden (* html) from non-IE browsers. */ | ||
| + | } | ||
| + | * html div.MenuBar ul li table, | ||
| + | * html div.MenuBar ul li table td { | ||
| + | border: 0; /* Required for IE55 and IE6. | ||
| + | Hidden (* html) from non-IE browsers. */ | ||
| + | } | ||
| + | /*------------------------------------------------------------------------------- Default Colors */ | ||
| + | div.MenuBar { | ||
| + | background-color: Menu; | ||
| + | border-bottom: 1px solid ButtonShadow; | ||
| + | } | ||
| + | div.MenuBar a { | ||
| + | background-color: Menu; /* Top-level unselected items. */ | ||
| + | color: MenuText; | ||
| + | } | ||
| + | div.MenuBar ul li:hover a, | ||
| + | div.MenuBar ul li a:hover { | ||
| + | color: #ea7f16; | ||
| + | background-color: Highlight; /* Top-level selected item. */ | ||
| + | color: HighlightText; | ||
| + | } | ||
| + | /*...............................................................................................*/ | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu { | ||
| + | background-color: ButtonShadow; /* Sets the drop-down menu "effective border" color. */ | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a { | ||
| + | background-color: Menu; Drop-down menu unselected items. | ||
| + | Sets the drop-down menu "effective background" color. */ | ||
| + | color: MenuText; | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover { | ||
| + | background-color: Highlight; /* Drop-down menu selected item. */ | ||
| + | color: HighlightText; | ||
| + | } | ||
| + | /*...............................................................................................*/ | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu { | ||
| + | background-color: ButtonShadow; /* Sets the side menu "effective border" color. */ | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu li a { | ||
| + | background-color: Menu; /* Side menu unselected items. | ||
| + | Sets the side menu "effective background" color. */ | ||
| + | color: MenuText; | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu li a:hover, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu li a:hover { | ||
| + | background-color: Highlight; /* Side menu selected item. */ | ||
| + | color: HighlightText; | ||
| + | } | ||
| + | /*-----------------------------------------------------------------------------------------------*/ | ||
| + | /*Script-Free 3-Level Menu 1.2 Tailor | ||
| + | www.CesarDaniel.info | ||
| + | /*-------------------------------------------------------------------------------------- General */ | ||
| + | body { | ||
| + | background: white; | ||
| + | color: black; | ||
| + | margin: 0; | ||
| + | border: 0; | ||
| + | padding: 0; | ||
| + | } | ||
| + | |||
| + | |||
| + | div.MenuBar { | ||
| + | font: 13px arial, helvetica, sans-serif; | ||
| + | } | ||
| + | div.MenuBar ul { | ||
| + | font: 13px arial, helvetica, sans-serif; /* Required for IE55. */ | ||
| + | } | ||
| + | /*--------------------------------------------------------------------------------------- Colors */ | ||
| + | div.MenuBar { | ||
| + | background-color: black; /* Selected item. */ | ||
| + | color: #FFFFFF; | ||
| + | border-bottom: 1px solid ButtonShadow; | ||
| + | } | ||
| + | div.MenuBar a, | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a, | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu li a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu li a { | ||
| + | background-color: black; /* Selected item. */ | ||
| + | color: #FFFFFF; | ||
| + | } | ||
| + | div.MenuBar ul li:hover a, | ||
| + | div.MenuBar ul li a:hover, | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover a, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover, | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu li a:hover, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu li a:hover { | ||
| + | background-color: #FF0000; /* Selected item. */ | ||
| + | color: #FFFFFF; | ||
| + | } | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu, | ||
| + | div.MenuBar ul li:hover ul.DropDownMenu li:hover ul.SideMenu, | ||
| + | div.MenuBar ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu { | ||
| + | background-color: ButtonShadow; /* Sets the drop-down and side menus "effective border" color. */ | ||
| + | } | ||
| + | /*--------------------------------------------------------------------------------------- Widths */ | ||
| + | /* | ||
| + | |||
| + | /* | ||
| + | Menu Bar 1 | ||
| + | Drop-Down Menu #2 | ||
| + | */ | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu#MB1-DDM4, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu#MB1-DDM4, | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu#MB1-DDM4 li a, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu#MB1-DDM4 li a { | ||
| + | width: 11em; /* Drop-down menu width. */ | ||
| + | } | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu#MB1-DDM5, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu#MB1-DDM5, | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu#MB1-DDM5 li a, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu#MB1-DDM5 li a { | ||
| + | width: 12em; /* Drop-down menu width. */ | ||
| + | } | ||
| + | |||
| + | /*...............................................................................................*/ | ||
| + | /* | ||
| + | Menu Bar 1 | ||
| + | Drop-Down Menu #2 | ||
| + | Side Menu #1 | ||
| + | */ | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM1, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM1 { | ||
| + | left: 15.5em !important; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. */ | ||
| + | } | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM1, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM1, | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM1 li a, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM1 li a { | ||
| + | width: 10em; /* Side menu width. */ | ||
| + | } | ||
| + | /*...............................................................................................*/ | ||
| + | /* | ||
| + | Menu Bar 1 | ||
| + | Drop-Down Menu #2 | ||
| + | Side Menu #2 | ||
| + | */ | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM2, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM2 { | ||
| + | left: 15.5em !important; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. */ | ||
| + | } | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM2, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM2, | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM2 li a, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM2 li a { | ||
| + | width: 10em; /* Side menu width. */ | ||
| + | } | ||
| + | /*...............................................................................................*/ | ||
| + | /* | ||
| + | Menu Bar 1 | ||
| + | Drop-Down Menu #2 | ||
| + | Side Menu #3 | ||
| + | */ | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM3, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM3 { | ||
| + | left: 15.5em !important; /* Places the side menu to the right of the drop-down menu. | ||
| + | Keep it sync with the drop-down menu width. */ | ||
| + | } | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM3, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM3, | ||
| + | div.MenuBar#navi ul li:hover ul.DropDownMenu li:hover ul.SideMenu#MB1-DDM2-SM3 li a, | ||
| + | div.MenuBar#navi ul li a:hover ul.DropDownMenu li a:hover ul.SideMenu#MB1-DDM2-SM3 li a { | ||
| + | width: 10em; /* Side menu width. */ | ||
| + | } | ||
| + | /*...............................................................................................*/ | ||
| + | |||
| + | </style> | ||
| + | |||
| + | |||
| + | <body> | ||
| + | <div id="header"><img src="https://static.igem.org/mediawiki/2010/3/31/RMIT_logo.png" /></div> | ||
| + | <div class='MenuBar' id="navi"> | ||
| + | <ul> | ||
| + | <li> | ||
| + | <a href="https://2010.igem.org/Team:RMIT_Australia" style="color: white">Home | ||
| + | <!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2010.igem.org/Team:RMIT_Australia/Team" style="color: white">Team | ||
| + | <!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2010.igem.org/Team:RMIT_Australia/Project" style="color: white">Projects | ||
| + | <!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | |||
| + | </li> | ||
| + | <li> | ||
| + | |||
| + | <a href="https://2010.igem.org/Team:RMIT_Australia/Parts" style="color: white">Parts | ||
| + | <!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2010.igem.org/Team:RMIT_Australia/Modeling" style="color:white">Modelling | ||
| + | <!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2010.igem.org/Team:RMIT_Australia/Notebook" style="color: white">Notebook | ||
| + | <!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | |||
| + | |||
| + | </li> | ||
| + | <li> | ||
| + | <a href="https://2010.igem.org/Team:RMIT_Australia/Safety" style="color: white">Safety | ||
| + | <!--[if gt IE 6]><!--></a><!--<![endif]--> | ||
| + | <!--[if lt IE 7]><table border="0" cellpadding="0" cellspacing="0"><tr><td><![endif]--> | ||
| + | <!--[if lte IE 6]></td></tr></table></a><![endif]--> | ||
| + | </li> | ||
| + | |||
| + | <li> | ||
| + | <a href="https://2010.igem.org/Team:RMIT_Australia/Sponsors" style="color: white">Sponsors</a> | ||
| + | </li> | ||
| + | </ul> | ||
| + | |||
</div> | </div> | ||
| + | <div id="header_bottom"><img src="https://static.igem.org/mediawiki/2008/e/ef/Navi_bg.gif" alt="" / ></div> | ||
| + | </body> | ||
</html> | </html> | ||
| - | |||
| - | + | = '''The Project''' = | |
| - | + | <p> | |
| - | + | The RMIT University iGEM project will attempt to create a peptide expression platform using Synthetic Biology. The recombinant production of protein is routine in science and industry. Despite protein expression being achievable, peptide expression is frequently too complicated or not economically viable; peptides are small and often contain very little – or even no secondary structure – making them highly susceptible to proteolysis and other cellular processes. | |
| - | + | </p><p> | |
| - | + | In the laboratory for research purposes, carrier proteins with affinity for a purification matrix are commonly employed, allowing “protection” of the peptide intracellularly and providing a defined bioprocess. There are several limitations to this technology that curtails the scale: Firstly, the process is extremely inefficient as it expends the feedstock and cellular metabolism to essentially produce large amounts of a worthless carrier protein (as much as 98% of the protein-peptide fused product by mass)., Secondly, affinity resins used to capture the carrier protein are expensive and rarely congenial to scalable chromatography unit operations such as expanded bed chromatography., Thirdly, there is a requirement to separate the peptide from the carrier protein that often utilises a protease. Proteases are expensive at elevated scales, and despite popular beliefs are actually inefficient with best estimates ranging in ratio yields of 1:40 – 1:200 for the desired product as a “good” outcome due to inefficiencies, non-specific cleavage or loss during purification. For these reasons there are very few commodity peptides made recombinantly or even by semi-synthetic methods, with solid-phase peptide synthesis remaining the industry benchmark. | |
| - | + | </p><p> | |
| - | + | The RMIT University iGEM project poses the question: Can the bottlenecks of purification and proteolysis be removed from the workflow to economically produce a peptide and commodity carrier protein? In this project we propose the thermostable protein Taq polymerase may be used as a carrier molecule to allow for the production of a peptide that will be attached to the Taq commodity enzyme. The peptide-Taq fusion will be attached by a thermolabile bond – an Asp/Pro bond in between the peptide and the Taq Polymerase, with the intention of releasing the peptide-Taq fusion products by thermolysis. The hypothetical bioprocess will involve heating the bacteria below the threshold of the thermolabile bond, then following clarification or polishing, heating above the threshold to separate the peptide from the Taq protein. The peptide and Taq may then be purified allowing two income streams from the manufacture. | |
| - | + | </p><p> | |
| - | + | Our project seeks to produce several unique parts for the registry: Firstly, a plasmid and system for producing Taq polymerase as a useful part for future iGEM projects undertaking PCR as part of their experiments., Secondly, an expression device will be provided to the registry that will allow peptides to be inserted easily and quickly by a process of ligand independent cloning into the device to produce any peptide at will, or even allow the rapid and economical production of a library of peptides for drug design and screening. | |
| - | + | ||
| - | + | = '''Project Details''' = | |
| + | |||
| + | |||
| + | |||
| + | == Creation of the Taq Polymerase Part == | ||
| + | |||
| + | <br> | ||
| + | Taq polymerase was chosen as a carrier molecule because of it's physico-chemical properties - most notably, Taq's thermostability at high temperatures; being able to fold and unfold repeatedly and through temperature gradients up to 95degC. Taq polymerase is a DNA-dependent DNA polymerase, using any DNA strand as a substrate and, conceivably, Taq expression levels within E.coli will be limited by toxic effects the protein will have when binding to genomic DNA and disrupting cellular functions. Fortunately, the natural polymerase function is not required for this project and so mutagenesis was chosen to knock-out DNA-binding, but retaining the favourable physico-chemical properties of Taq. | ||
| - | + | Using the ExPASy proteomics server the aliphatic index, instability index, extinction coefficient and theoretical isoelectric point were determined to distinguish mutations that increase the thermostability and through literature research, amino acids which are essential for catalytic activity of Taq were identified and these mutations to inactivate polymerase activity and reduce DNA binding were chosen: N485V, S515L, K540L, R659I and K663L. Mutating these residues would render Taq incapable of polymerase activity but retain thermostability. Mutations to enable ligation independent cloning (LIC) were added to the N and the C terminus (see LIC section for more information), along with the prefix(P) and the suffix(S). These mutation were incorporated into Taq sequence to produce the mutTaq sequence (see modeling for more information). The sequence was then sent to Mr. Gene (Gene Art) for the production of the (pMA-RQ) vector containing the muTAQ sequence conforming to standard 10 cloning (below is a pictorial representation of the vector containing mutTaq). | |
| + | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | [[Image:gav.jpg|400px|centre]] | ||
| + | <br> | ||
| + | == Making a Controlled T7 Expression Vector == | ||
| + | <br> | ||
| + | The RMIT2010 team is interested in producing a protein expression vector for producing peptides fused with Taq. Arguably the best protein expression systems are the pET vectors. These vector systems use a T7 promoter regulated by the Lac Operon elements. This means that: | ||
| + | * The LacO/LacI binding site is a repressor site, inhibiting the RNA polymerase to transcribe the mRNA. By adding '''Lactose''' or its analog '''IPTG''', the structure of the LacI repressor is changes, thus not being able to bind to the LacO site. this allows transcription to occur. | ||
| - | + | * Another regulatory element in the Lac system is the CAP binding site, which is located just upstream of the promoter region. Its role is to enhance transcription of the operon. The CAP protein is found throughout the cell in its inactive form - being activated by the molecule cAMP. When the levels of cAMP are high, there is more active CAP protein in the cell. Interestingly enough, the levels of cAMP are inversely proportional to the levels of glucose in the cell, so by depriving the cell from this carbon source the levels of transcription will thrive. | |
| - | + | The T7 sequence is very small and though several designs of Lac/T7 systems have been designed in the parts registry, none have been made available in the distribution. When cloning T7 components, the small sequence means that there is large difficulty in gel electrophoresis detection and there is ambiguity as to whether a T7 promoter will work in the context of some promoter designs. Our T7 part is based on the iGEM known part BBa_J04500 which is a functional Lac operon promoter (plus RBS) that has been benchmarked and tested in several iGEM projects, therefore providing a strong controllable and usable part for future iGEM distributions. | |
| + | This part - as previously stated - was made from the part BBa_J04500. The first challenge that we encountered was to remove the current promoter elements found in the part: -35 and -10 elements. This was overcome using Quikchange XL Site-directed Mutagenesis to introduce unique restriction sites to allow the removal of the promoter region. The -35 element was mutated to include an Afl''II'' restriction site and the -10 element was mutated to a Cla''I'' site. The T7 promoter was inserted via primers with ends to match the restriction sites, creating a new part when ligated into iGEM standard plasmid pSB1C3. | ||
| + | Part construction was confirmed through restriction digest and sequencing and is compatible with all assembly standards. This part is roughly 220bp - an appreciable size difference to observe in gel electrophoresis when undertaking assembly. | ||
| + | <br> | ||
| + | <br> | ||
| + | [[Image:T7part.png|700px|centre]] | ||
| + | <br> | ||
| + | <br> | ||
| + | == Ligation Independent Cloning == | ||
| + | Ligation independent cloning (LIC) is a new procedure that exploits the 3’-5’ exonuclease activity of DNA polymerases to produce sequence-specific overhangs for target primer insertion. In the LIC procedure, the DNA is cut with a restriction enzyme(s) and DNA polymerase is added in the presence of only one dNTP. In the absence of all other dNTPs, the equilibrium of the polymerase activity shift to exonuclease activity and nucleotides are removed from the point of the restriction digest until the polymerase encounters a nucleotide that corresponds to the single dNTP present. At this point the exonulease activity stops and is counteracted by the 5’-3’ polymerisation activity which prevents further changes to the DNA due to only one dNTP being present. This results in a polymerase-digested vector with non-homologous sequences of sticky-ends. Primers can then be designed with complementarity for the sticky-ends, allowing for the insertion of a desired peptide sequence within the vector by simply annealing the primers to the vector and transformation into cells. | ||
| + | The thermostable protein Taq polymerase was chosen as a carrier molecule, to facilitate the production of a peptide as a peptide-Taq fusion protein separated by a thermolabile bond, such as an Asp/Pro bond. The proposed bioprocess would then involve heating the bacteria below the threshold of the thermolabile bond for simplified lysis, then following clarification or polishing, heating above the threshold of the thermolabile bond to separate the peptide from the Taq protein allowing use expression immediately in assays, or providing a simple starting source for final purification. | ||
| - | + | The peptide plasmid was also designed with the intention of allowing peptides to be inserted easily and quickly by a process of Ligation Independent Cloning (LIC) (refns) to produce a desired peptide at will, or even allow the rapid and economical production of a library of peptides for drug design and screening. Two test peptides were chosen to explore the capacity of the peptide expression platform being Losartan and arginine vasopressin. Arginine vasopressin and the angiotensin antagonist Losartan were chosen because they both had short amino acid sequences that we could design primers for. | |
| + | With Taq, it is feasible that the first and last 10 amino acids of Taq may be deleted without greatly influencing the 3D structure . The first 10 amino acids are not present in the original crystal structure and both the N- and C- termini are in regions which project a putative peptide more generally away from the protein structure, with relatively few intramolecular native contacts observable in this region. To further facilitate the separation and projection of expressed peptides away from the native protein structure, a hydrophilic spacer arm was also included in the design of mutTaq at the N- and C- termini consisting of a simple GSSG motif that produces a known extended conformation of ~9.88Å . | ||
| + | Because Taq has a DNA sequence with an unusually rich GC content it was determine that the appropriate dNTP to generate the specific overhangs should be either dATP or dTTP as this allows a lengthy digestion by a DNA polymerase of >19 nucleotides. The generation of these sticky ends would require digestion by the polymerase into the coding region for both the N- or C- terminus of the mutTaq but also into important expression features of the vector such as the stop codon, start codon and ribosomal binding site. An NdeI restriction site (CA^TATG) was therefore designed at the start codon of mutTaq (5’ coding region) and nucleotides for a GSSG spacer arm and AvrII restriction site (C^CTAGG) were designed at the 3’ coding region. The NdeI and AvrII restriction enzymes were considered favourable restriction enzymes to employ for process control and flexibility as they can be heat inactivated at 65ºC and 80ºC respectively, have more broad buffer compatibilities, are not sensitive to methylation (dam, dcm, CpG) and are suitable for extended digestion (e.g. >8h or overnight). | ||
| + | <br> | ||
| + | ===Design of Mutated Taq Plasmid for LIC=== | ||
| + | [[Image:LIC1.png|800px]] | ||
| - | |||
| + | ===Design of Mutated Taq Plasmid after T4 Polymerase Exonuclease Activity=== | ||
| + | [[Image:LIC2.png|800px]] | ||
| - | |||
| + | ===Design of Mutated Taq Plasmid after LIC with Inserted Peptide=== | ||
| + | [[Image:LIC3.png|800px]] | ||
| - | == | + | == Peptides == |
| + | ===Angiotensin-Losartan=== | ||
| + | The Angiotensin polypeptide causes vasoconstriction, raises blood pressure and causes the release of aldosterone from the adrenal cortex. It is formed by the action of renin on angiotensinogen, an alpha-2-glycoprotein that is produced in the liver and circulates in the blood. Renin acts as an enzyme in response to lowered blood pressure volume in the conversion of angiotensinogen to angiotensin I which is then rapidly hydrolysed to form the active compound angiotensin II. This renin-angiotensin system also regulates electrolyte balance and arterial blood pressure. ACE (angiotensin -converting enzyme) is a dipeptidyl carboxypeptidase that catalyses the conversion of angiotensin I to angiotensin II by removing the two terminal amino acids. ACE inhibiting agents are used in hypertension control and for the protection of kidneys in diabetes mellitus. | ||
| + | Angiotensin receptor blocker (ARB) such as Losartan (a peptide we desired to make in this project) is a class of drugs acting on the renin –angiotensin-aldosterone system to block the action of angiotensin II on the AT1 receptors and thereby cause vasodilation (AT1 antagonist for the treatment of hypertension). Losartan is an oral drug shown to be the most effective therapeutic for treating hypertension. It is also a drug that is currently being considered as a treatment for Marfan’s syndrome. AT1 receptors are found in many tissues including vascular smooth muscle, adrenal gland, kidneys and heart. The RMIT IGEM team intend to anneal a short peptide-encoding primer into a bacterial backbone plasmid to produce Losartan for a fraction of the price needed currently to produce the drug commercially. The peptide sequence we intend on using is composed of N terminal-Asp-Arg-Val-Tyr-Val/IIe –His-Pro-Phe-C terminal. | ||
| + | ===Arginine Vasopressin=== | ||
| + | The second peptide we were interested in producing was arginine vasopressin which is an antidiuretic hormone secreted from the posterior pituitary gland that exerts two important biological functions; (i) potent vasoconstriction and (ii) as the main regulator of water homeostasis and extracellular fluid volume. The later antidiuretic properties of arginine vasopressin are the most biologically important, exerting therapeutic effects in the kidney and the vascular system and therefore being widely used in the treatment of diabetes insipidus. | ||
| + | G- coupled protein receptor (GPCR) agonists such as vasopressin, account for 40% of prescription pharmaceuticals on the market and are the core of medicine as GPCRs are the most investigated target sites in the pharmaceutical industry. | ||
| + | For these reasons and for its simplicity to assay, it has been chosen as a candidate to test the efficacy of the new technology. | ||
| + | Calcium ion assays are used to detect the activation of GCPRs as they are naturally produced upon activation of GPCRs coupled to Gq, the G protein involved in the activation of the arginine vasopressin receptor 1 that causes vasoconstriction. Increased intracellular calcium levels infer that the peptide has been produced and expressed. | ||
| + | Screening for adenosine 3':5'-cyclic monophosphate (cAMP), a second messenger produced when the receptor is activated, indicates the presence of a peptide such as vasopressin that has triggered this activation with the coupling of G proteins. Luminescent tags bind to the cAMP to allow for its detection. | ||
| - | + | <br> | |
Latest revision as of 00:41, 28 October 2010


The Project
The RMIT University iGEM project will attempt to create a peptide expression platform using Synthetic Biology. The recombinant production of protein is routine in science and industry. Despite protein expression being achievable, peptide expression is frequently too complicated or not economically viable; peptides are small and often contain very little – or even no secondary structure – making them highly susceptible to proteolysis and other cellular processes.
In the laboratory for research purposes, carrier proteins with affinity for a purification matrix are commonly employed, allowing “protection” of the peptide intracellularly and providing a defined bioprocess. There are several limitations to this technology that curtails the scale: Firstly, the process is extremely inefficient as it expends the feedstock and cellular metabolism to essentially produce large amounts of a worthless carrier protein (as much as 98% of the protein-peptide fused product by mass)., Secondly, affinity resins used to capture the carrier protein are expensive and rarely congenial to scalable chromatography unit operations such as expanded bed chromatography., Thirdly, there is a requirement to separate the peptide from the carrier protein that often utilises a protease. Proteases are expensive at elevated scales, and despite popular beliefs are actually inefficient with best estimates ranging in ratio yields of 1:40 – 1:200 for the desired product as a “good” outcome due to inefficiencies, non-specific cleavage or loss during purification. For these reasons there are very few commodity peptides made recombinantly or even by semi-synthetic methods, with solid-phase peptide synthesis remaining the industry benchmark.
The RMIT University iGEM project poses the question: Can the bottlenecks of purification and proteolysis be removed from the workflow to economically produce a peptide and commodity carrier protein? In this project we propose the thermostable protein Taq polymerase may be used as a carrier molecule to allow for the production of a peptide that will be attached to the Taq commodity enzyme. The peptide-Taq fusion will be attached by a thermolabile bond – an Asp/Pro bond in between the peptide and the Taq Polymerase, with the intention of releasing the peptide-Taq fusion products by thermolysis. The hypothetical bioprocess will involve heating the bacteria below the threshold of the thermolabile bond, then following clarification or polishing, heating above the threshold to separate the peptide from the Taq protein. The peptide and Taq may then be purified allowing two income streams from the manufacture.
Our project seeks to produce several unique parts for the registry: Firstly, a plasmid and system for producing Taq polymerase as a useful part for future iGEM projects undertaking PCR as part of their experiments., Secondly, an expression device will be provided to the registry that will allow peptides to be inserted easily and quickly by a process of ligand independent cloning into the device to produce any peptide at will, or even allow the rapid and economical production of a library of peptides for drug design and screening.
Project Details
Creation of the Taq Polymerase Part
Taq polymerase was chosen as a carrier molecule because of it's physico-chemical properties - most notably, Taq's thermostability at high temperatures; being able to fold and unfold repeatedly and through temperature gradients up to 95degC. Taq polymerase is a DNA-dependent DNA polymerase, using any DNA strand as a substrate and, conceivably, Taq expression levels within E.coli will be limited by toxic effects the protein will have when binding to genomic DNA and disrupting cellular functions. Fortunately, the natural polymerase function is not required for this project and so mutagenesis was chosen to knock-out DNA-binding, but retaining the favourable physico-chemical properties of Taq.
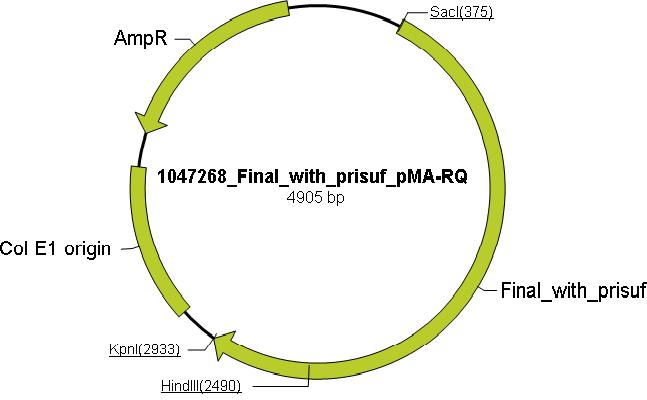
Using the ExPASy proteomics server the aliphatic index, instability index, extinction coefficient and theoretical isoelectric point were determined to distinguish mutations that increase the thermostability and through literature research, amino acids which are essential for catalytic activity of Taq were identified and these mutations to inactivate polymerase activity and reduce DNA binding were chosen: N485V, S515L, K540L, R659I and K663L. Mutating these residues would render Taq incapable of polymerase activity but retain thermostability. Mutations to enable ligation independent cloning (LIC) were added to the N and the C terminus (see LIC section for more information), along with the prefix(P) and the suffix(S). These mutation were incorporated into Taq sequence to produce the mutTaq sequence (see modeling for more information). The sequence was then sent to Mr. Gene (Gene Art) for the production of the (pMA-RQ) vector containing the muTAQ sequence conforming to standard 10 cloning (below is a pictorial representation of the vector containing mutTaq).
Making a Controlled T7 Expression Vector
The RMIT2010 team is interested in producing a protein expression vector for producing peptides fused with Taq. Arguably the best protein expression systems are the pET vectors. These vector systems use a T7 promoter regulated by the Lac Operon elements. This means that:
- The LacO/LacI binding site is a repressor site, inhibiting the RNA polymerase to transcribe the mRNA. By adding Lactose or its analog IPTG, the structure of the LacI repressor is changes, thus not being able to bind to the LacO site. this allows transcription to occur.
- Another regulatory element in the Lac system is the CAP binding site, which is located just upstream of the promoter region. Its role is to enhance transcription of the operon. The CAP protein is found throughout the cell in its inactive form - being activated by the molecule cAMP. When the levels of cAMP are high, there is more active CAP protein in the cell. Interestingly enough, the levels of cAMP are inversely proportional to the levels of glucose in the cell, so by depriving the cell from this carbon source the levels of transcription will thrive.
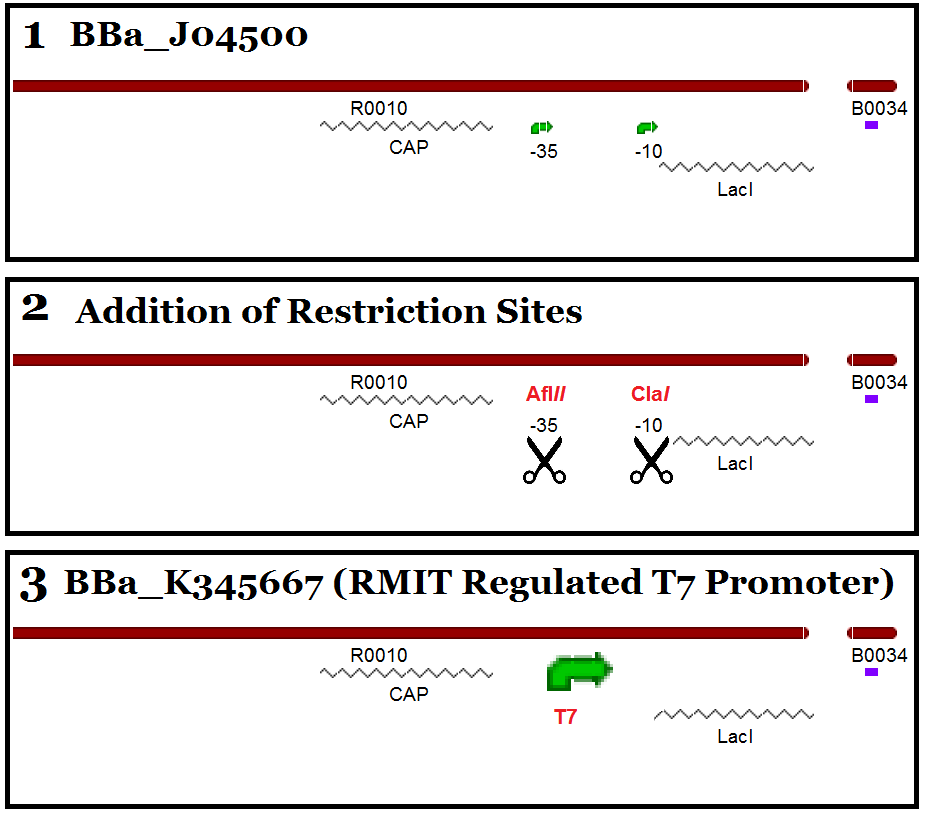
The T7 sequence is very small and though several designs of Lac/T7 systems have been designed in the parts registry, none have been made available in the distribution. When cloning T7 components, the small sequence means that there is large difficulty in gel electrophoresis detection and there is ambiguity as to whether a T7 promoter will work in the context of some promoter designs. Our T7 part is based on the iGEM known part BBa_J04500 which is a functional Lac operon promoter (plus RBS) that has been benchmarked and tested in several iGEM projects, therefore providing a strong controllable and usable part for future iGEM distributions.
This part - as previously stated - was made from the part BBa_J04500. The first challenge that we encountered was to remove the current promoter elements found in the part: -35 and -10 elements. This was overcome using Quikchange XL Site-directed Mutagenesis to introduce unique restriction sites to allow the removal of the promoter region. The -35 element was mutated to include an AflII restriction site and the -10 element was mutated to a ClaI site. The T7 promoter was inserted via primers with ends to match the restriction sites, creating a new part when ligated into iGEM standard plasmid pSB1C3.
Part construction was confirmed through restriction digest and sequencing and is compatible with all assembly standards. This part is roughly 220bp - an appreciable size difference to observe in gel electrophoresis when undertaking assembly.
Ligation Independent Cloning
Ligation independent cloning (LIC) is a new procedure that exploits the 3’-5’ exonuclease activity of DNA polymerases to produce sequence-specific overhangs for target primer insertion. In the LIC procedure, the DNA is cut with a restriction enzyme(s) and DNA polymerase is added in the presence of only one dNTP. In the absence of all other dNTPs, the equilibrium of the polymerase activity shift to exonuclease activity and nucleotides are removed from the point of the restriction digest until the polymerase encounters a nucleotide that corresponds to the single dNTP present. At this point the exonulease activity stops and is counteracted by the 5’-3’ polymerisation activity which prevents further changes to the DNA due to only one dNTP being present. This results in a polymerase-digested vector with non-homologous sequences of sticky-ends. Primers can then be designed with complementarity for the sticky-ends, allowing for the insertion of a desired peptide sequence within the vector by simply annealing the primers to the vector and transformation into cells.
The thermostable protein Taq polymerase was chosen as a carrier molecule, to facilitate the production of a peptide as a peptide-Taq fusion protein separated by a thermolabile bond, such as an Asp/Pro bond. The proposed bioprocess would then involve heating the bacteria below the threshold of the thermolabile bond for simplified lysis, then following clarification or polishing, heating above the threshold of the thermolabile bond to separate the peptide from the Taq protein allowing use expression immediately in assays, or providing a simple starting source for final purification.
The peptide plasmid was also designed with the intention of allowing peptides to be inserted easily and quickly by a process of Ligation Independent Cloning (LIC) (refns) to produce a desired peptide at will, or even allow the rapid and economical production of a library of peptides for drug design and screening. Two test peptides were chosen to explore the capacity of the peptide expression platform being Losartan and arginine vasopressin. Arginine vasopressin and the angiotensin antagonist Losartan were chosen because they both had short amino acid sequences that we could design primers for.
With Taq, it is feasible that the first and last 10 amino acids of Taq may be deleted without greatly influencing the 3D structure . The first 10 amino acids are not present in the original crystal structure and both the N- and C- termini are in regions which project a putative peptide more generally away from the protein structure, with relatively few intramolecular native contacts observable in this region. To further facilitate the separation and projection of expressed peptides away from the native protein structure, a hydrophilic spacer arm was also included in the design of mutTaq at the N- and C- termini consisting of a simple GSSG motif that produces a known extended conformation of ~9.88Å .
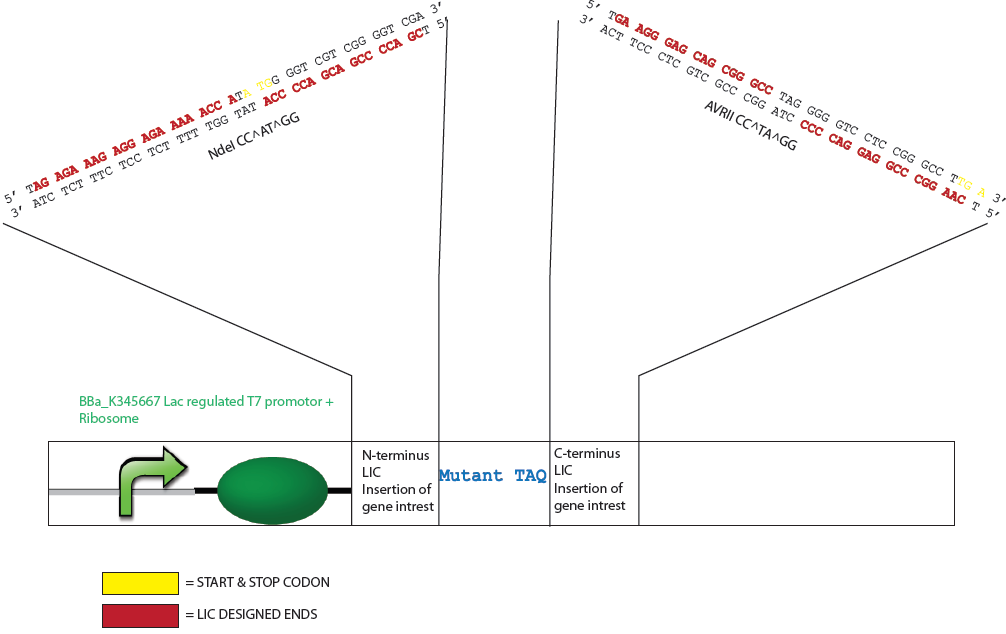
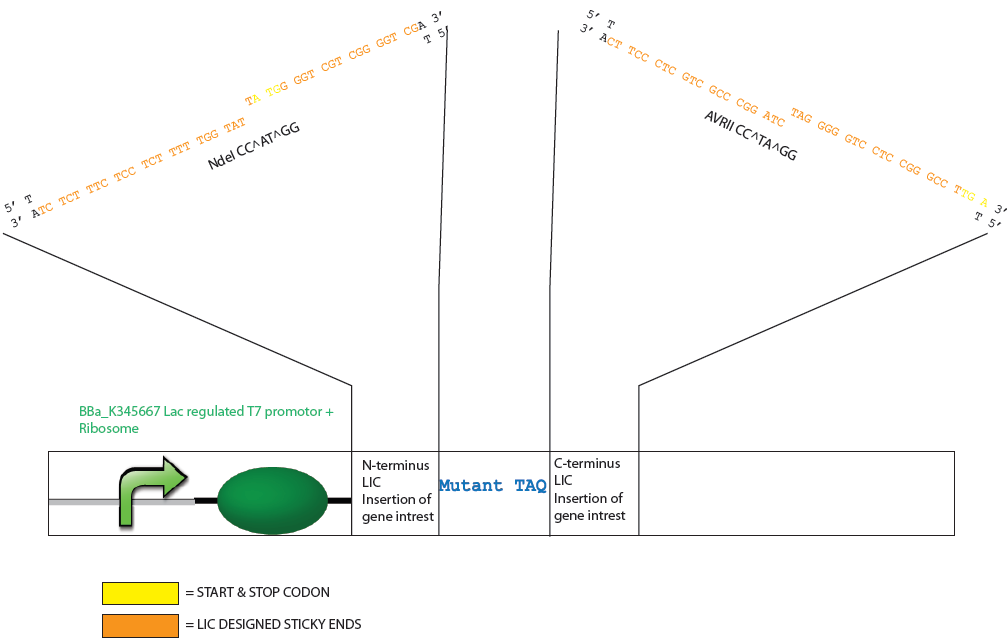
Because Taq has a DNA sequence with an unusually rich GC content it was determine that the appropriate dNTP to generate the specific overhangs should be either dATP or dTTP as this allows a lengthy digestion by a DNA polymerase of >19 nucleotides. The generation of these sticky ends would require digestion by the polymerase into the coding region for both the N- or C- terminus of the mutTaq but also into important expression features of the vector such as the stop codon, start codon and ribosomal binding site. An NdeI restriction site (CA^TATG) was therefore designed at the start codon of mutTaq (5’ coding region) and nucleotides for a GSSG spacer arm and AvrII restriction site (C^CTAGG) were designed at the 3’ coding region. The NdeI and AvrII restriction enzymes were considered favourable restriction enzymes to employ for process control and flexibility as they can be heat inactivated at 65ºC and 80ºC respectively, have more broad buffer compatibilities, are not sensitive to methylation (dam, dcm, CpG) and are suitable for extended digestion (e.g. >8h or overnight).
Design of Mutated Taq Plasmid for LIC
Design of Mutated Taq Plasmid after T4 Polymerase Exonuclease Activity
Design of Mutated Taq Plasmid after LIC with Inserted Peptide
Peptides
Angiotensin-Losartan
The Angiotensin polypeptide causes vasoconstriction, raises blood pressure and causes the release of aldosterone from the adrenal cortex. It is formed by the action of renin on angiotensinogen, an alpha-2-glycoprotein that is produced in the liver and circulates in the blood. Renin acts as an enzyme in response to lowered blood pressure volume in the conversion of angiotensinogen to angiotensin I which is then rapidly hydrolysed to form the active compound angiotensin II. This renin-angiotensin system also regulates electrolyte balance and arterial blood pressure. ACE (angiotensin -converting enzyme) is a dipeptidyl carboxypeptidase that catalyses the conversion of angiotensin I to angiotensin II by removing the two terminal amino acids. ACE inhibiting agents are used in hypertension control and for the protection of kidneys in diabetes mellitus.
Angiotensin receptor blocker (ARB) such as Losartan (a peptide we desired to make in this project) is a class of drugs acting on the renin –angiotensin-aldosterone system to block the action of angiotensin II on the AT1 receptors and thereby cause vasodilation (AT1 antagonist for the treatment of hypertension). Losartan is an oral drug shown to be the most effective therapeutic for treating hypertension. It is also a drug that is currently being considered as a treatment for Marfan’s syndrome. AT1 receptors are found in many tissues including vascular smooth muscle, adrenal gland, kidneys and heart. The RMIT IGEM team intend to anneal a short peptide-encoding primer into a bacterial backbone plasmid to produce Losartan for a fraction of the price needed currently to produce the drug commercially. The peptide sequence we intend on using is composed of N terminal-Asp-Arg-Val-Tyr-Val/IIe –His-Pro-Phe-C terminal.
Arginine Vasopressin
The second peptide we were interested in producing was arginine vasopressin which is an antidiuretic hormone secreted from the posterior pituitary gland that exerts two important biological functions; (i) potent vasoconstriction and (ii) as the main regulator of water homeostasis and extracellular fluid volume. The later antidiuretic properties of arginine vasopressin are the most biologically important, exerting therapeutic effects in the kidney and the vascular system and therefore being widely used in the treatment of diabetes insipidus. G- coupled protein receptor (GPCR) agonists such as vasopressin, account for 40% of prescription pharmaceuticals on the market and are the core of medicine as GPCRs are the most investigated target sites in the pharmaceutical industry. For these reasons and for its simplicity to assay, it has been chosen as a candidate to test the efficacy of the new technology. Calcium ion assays are used to detect the activation of GCPRs as they are naturally produced upon activation of GPCRs coupled to Gq, the G protein involved in the activation of the arginine vasopressin receptor 1 that causes vasoconstriction. Increased intracellular calcium levels infer that the peptide has been produced and expressed. Screening for adenosine 3':5'-cyclic monophosphate (cAMP), a second messenger produced when the receptor is activated, indicates the presence of a peptide such as vasopressin that has triggered this activation with the coupling of G proteins. Luminescent tags bind to the cAMP to allow for its detection.
 "
"