Team:Peking/Project/Biosensor/Bioreporter
Traffic-Light Bioassay For Application Ease
Traditional bioassays with biosensor bacteria are usually calibrated with analyte solutions of known concentrations that are analyzed along with the samples of interest (Chakraborty et al., 2008; Hansen and Sorensen, 2000). This is done as bioreporter output (fluorescence or colour) does not only depend on the target concentration, but also on the incubation time and physiological activity of the cells in the assay. Comparing the biosensor output with standardized colour tables in the field application seems rather difficult and error-prone.
To solve this hard truth, a new approach called ‘traffic light’ heavy metal bioassay was then developed. Previous work has shown that the “traffic light” bioassay could work independently of external calibration of the bioreporter output, thus to control assay variations and to improve application ease(Anke Wackwitz, 2008). Actually, an internal calibration based on the use of multiple isogenic bioreporter cell lines with the same output but drastically different sensitivity at a given heavy metal concentration is proceeded during this bioassy.

Fig 1. The traffic light bioassay uses, for example, four isogenic strains with the same reporter output but differing in the compound concentration threshold at which their reporter circuit is activated. The number of reporter strains reacting to a sample (rather than their reporter signal intensity per se) is then representative for the compound concentration range. Adapted from (van der Meer and Belkin)
Specially speaking, we pyramided the information collected during the Promoter Characterization, Operation Characterization and Modeling. It has been shown that the expression level of MerR and the binding affinity of MerR to the cognate promoter dyad sequence both significantly determinate the bacterial sensitivity to mercury — higher MerR expression intensity and lower binding affinity of MerR to DNA target would result in a less sensitive bacterial mercury sensor and vice versa. In order to verify this, we constructed a reporter system similar to the ones we used before, as shown in Fig 2C.(to read more…).




Fig 2. Result of pyramiding. According to the information collected in promoter characterization and operation characterization, both the expression level of MerR and the semiconserved region of MerR binding site could determine the threshold of mercury sensitivity. (A) Each combination was carefully characterized. All of the dose response curves represented as a hill function. (B) When comparing these combinations, we can easily find that both of the factors worked as expected: for instance, BBa_J23101+Mutant 3 represents a higher threshold than that of BBa_J23101+Mutant 88 and BBa_J23103+Mutant 88 has a higher threshold than BBa_J23101+Mutant 88. (C) The genetic construction of the system used for characterization.
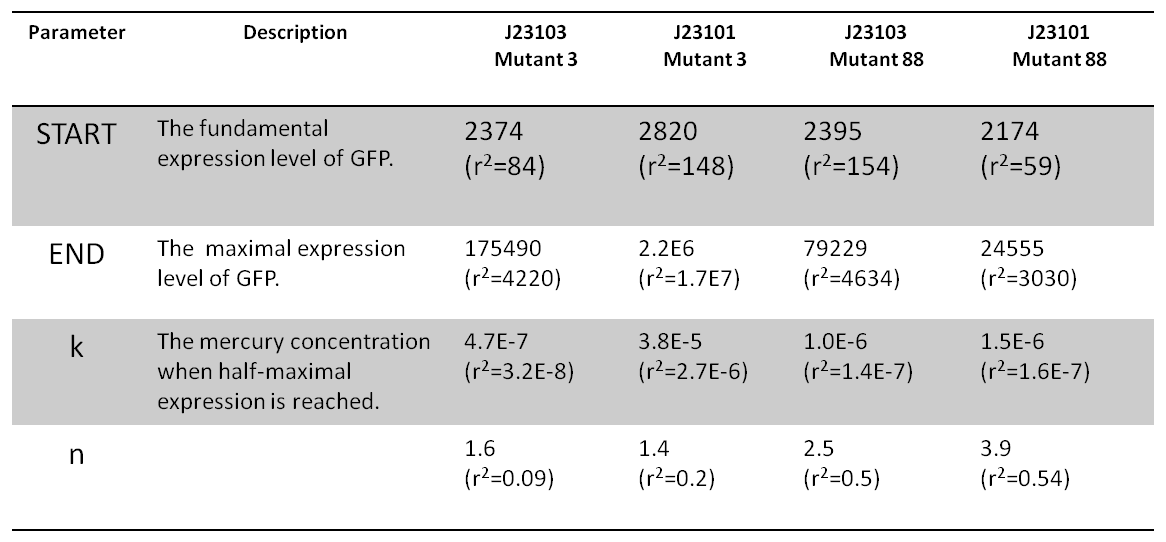
Table 2 The parameters of Hill function fitting.
Therefore, 4 isogenic biosensor strains with the same reporter output (beta-galactosidase) but differing in the mercury concentration threshold at which their reporter circuit is activated was constructed as is shown in Fig 3.
Interestingly, when we employed full length LacZ gene (BBa_I732017) as the reporter gene for design in Fig 3, a significant leakage expression emerged which was as high as we could not tell the difference in intensity of the beta-galactosidase activity between experimental group and the control. However, when using LacZ alpha fragment instead, we found that the leakage expression became negligible – Hg (II) induction would significantly activate the beta-galactosidase activity compared with the control and bacteria bearing the mercury sensing device behaved in a dose-response manner in response to mercury concentration gradient (Fig 4), which implied that when separated into 2 peptide fragments, the enzymatic activity of LacZ (beta-galactosidase) decreased remarkably and the decrement could be rescued by higher expression level of the alpha fragment. It was probably because the alpha-complementary process is mediated by intra-molecular non-covalent interactions. In comparison with the full length LacZ, alpha-complement forms the correct conformation for beta-galactosidase activity in lower possibility. Therefore, mercury sensing device exploiting full length LacZ as reporter gene can not actually sense the mercury. However, it may act as the positive control in the traffic light assay, which exhibits beta-galactosidase activity whether mercury is in presence or not (Fig 3).





Fig 3. Scheme of construction of 4 isogenic biosensor strains. It was a pyramiding process during which the phenotypes of bacterial reporter developed previously in our project were combined, in order to implement strains with different mercury sensitivity. (E) BBa_J23103, BBa_J23101 and BBa_J23117 belong to a constitutive promoter library from partsregistry, among which BBa_J23103 is a very weak one, BBa_J23117 is medium and BBa_J23101 is the second strongest in the library. (A)Expression of MerR driven by weak constitutive promoter BBa_J23103 on low copy number plasmid backbone pSB3K3 was pyramided with wild type PmerT which is the most mercury-sensitive promoter combined with LacZ alpha fragment as the reporter gene on plasmid backbone pSB1A3. It is an extreme to confer the bioreporter the most sensitivity, based on our previous results. (B) Strong promoter BBa_J23101 drive the high expression intensity of MerR on plasmid backbone pSB1A2, while PmerT 88 on pSB3K3 is the most insensitive mercury-response promoter developed before. Reporter gene is still lacZ alpha fragment. This is the other extreme of MerR expression intensity, aiming to endow the bacteria the most insensitivity. (C) Promoter BBa_J23117 is medium compared with other 2 constitutive promoters, and the same with PmerT 03 which was screened out previously. This mercury sensing device is expected to be more sensitive than (A) and less than (B). (D) This device is similar to (A), except the reporter gene was replaced by LacZ full length gene. As mentioned in the context, when acting as the reporter gene, full length LacZ represents a significantly leakage expression, giving output regardless of the input, of which we took the advantage to regard it as the positive control in traffic light bioassay.
With completion of 4 isogenic biosensor strain construction, traffic light bioassay was performed in 96-well plates to allow visual signal detection by digital imaging (Fig 4), rather than other detection methods which are time-comsuming or need costly instruments. For each biosensor strain the bioassay was performed in triplicate with 24 different mercury concentrations ranging from 10^-9 M to 10^-5 M.
fig 4
Results demonstrated that the bioreporter system using combinations of these biosensor strains to define mercury concentration ranges at which none, one or more biosensor strains gave qualitative (yes/no) visible signals that were relatively independent of incubation time or bioreporter activity. The discriminated concentration ranges would fit very well with the current permissive (e.g. World Health Organization) levels of mercury in common aquatic environment, such as drinking water.
Reference
Anke Wackwitz, H.H., Antonis Chatzinotas, Uta Breuer, Christelle Vogne, Jan Roelof Van Der Meer (2008). Internal arsenite bioassay calibration using multiple bioreporter cell lines. Microbial Biotechnology 1, 149-157.
Chakraborty, T., Babu, P.G., Alam, A., and Chaudhari, A. (2008). GFP expressing bacterial biosensor to measure lead contamination in aquatic environment. Current Science 94, 800-805.
Hansen, L.H., and Sorensen, S.J. (2000). Versatile biosensor vectors for detection and quantification of mercury. FEMS Microbiol Lett 193, 123-127.
van der Meer, J.R., and Belkin, S. Where microbiology meets microengineering: design and applications of reporter bacteria. Nat Rev Microbiol 8, 511-522.
 "
"