Team:Paris Liliane Bettencourt/Project/Population counter/Results
From 2010.igem.org
| Line 44: | Line 44: | ||
<br><br></html> | <br><br></html> | ||
| + | |||
==Proof that IntI1 integrase works !== | ==Proof that IntI1 integrase works !== | ||
| + | <html><p style="display:block;"> | ||
<br> Then we wanted to see if the IntI1 integrase actually works. We have done a double transformation "RFP Counter" (on pSB1A2 which is a high copy plasmid) + "pBAD-IntI1" (on pSulib which is a low copy plasmid with kanamycin resistance) to obtain a working counter. The protocol is quite simple : we dilute an overnight culture, wait until OD 0,2 and add arabinose to begin the test (t=0h). At regular time intervals, we plated on plates containing ampicillin (this is not important if the cells loose the "pBad-Int" plasmid because we don't want recombinations to happen on the plates). | <br> Then we wanted to see if the IntI1 integrase actually works. We have done a double transformation "RFP Counter" (on pSB1A2 which is a high copy plasmid) + "pBAD-IntI1" (on pSulib which is a low copy plasmid with kanamycin resistance) to obtain a working counter. The protocol is quite simple : we dilute an overnight culture, wait until OD 0,2 and add arabinose to begin the test (t=0h). At regular time intervals, we plated on plates containing ampicillin (this is not important if the cells loose the "pBad-Int" plasmid because we don't want recombinations to happen on the plates). | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<br> This experiment shows us that IntI1 is effective ! The double terminator flanked by attC sites has been excised and the constitutive promoter upstream permits the expression of RFP. However, we have 2 important notes : | <br> This experiment shows us that IntI1 is effective ! The double terminator flanked by attC sites has been excised and the constitutive promoter upstream permits the expression of RFP. However, we have 2 important notes : | ||
| - | * At the beginning of the test, there already colonies that are fluorescents... We can explain this fact by the leaking of pBad that controls the expression of the integrase. Thereby the integrase is expressed at a low level and catalyzes the excision of the terminator. To overcome this problem we have to add glucose to the media which blocks the pBAD promoter and avoid recombination events to occur . | + | </html> |
| + | * At the beginning of the test, there already colonies that are fluorescents... We can explain this fact by the leaking of pBad that controls the expression of the integrase. Thereby the integrase is expressed at a low level and catalyzes the excision of the terminator. To overcome this problem we have to add glucose to the media which blocks the pBAD promoter and avoid recombination events to occur. | ||
*The expression of RFP is inhomogeneous because the counter is on a high copy plasmid. So some cells have 2 recombined plasmids while others have 20 recombined plasmids ! To obtain a more linear response we have integrated our counter in a low copy plasmid (pSB4A5). In this way, there is more chance that only one event will happen in each cell. | *The expression of RFP is inhomogeneous because the counter is on a high copy plasmid. So some cells have 2 recombined plasmids while others have 20 recombined plasmids ! To obtain a more linear response we have integrated our counter in a low copy plasmid (pSB4A5). In this way, there is more chance that only one event will happen in each cell. | ||
<br><br> | <br><br> | ||
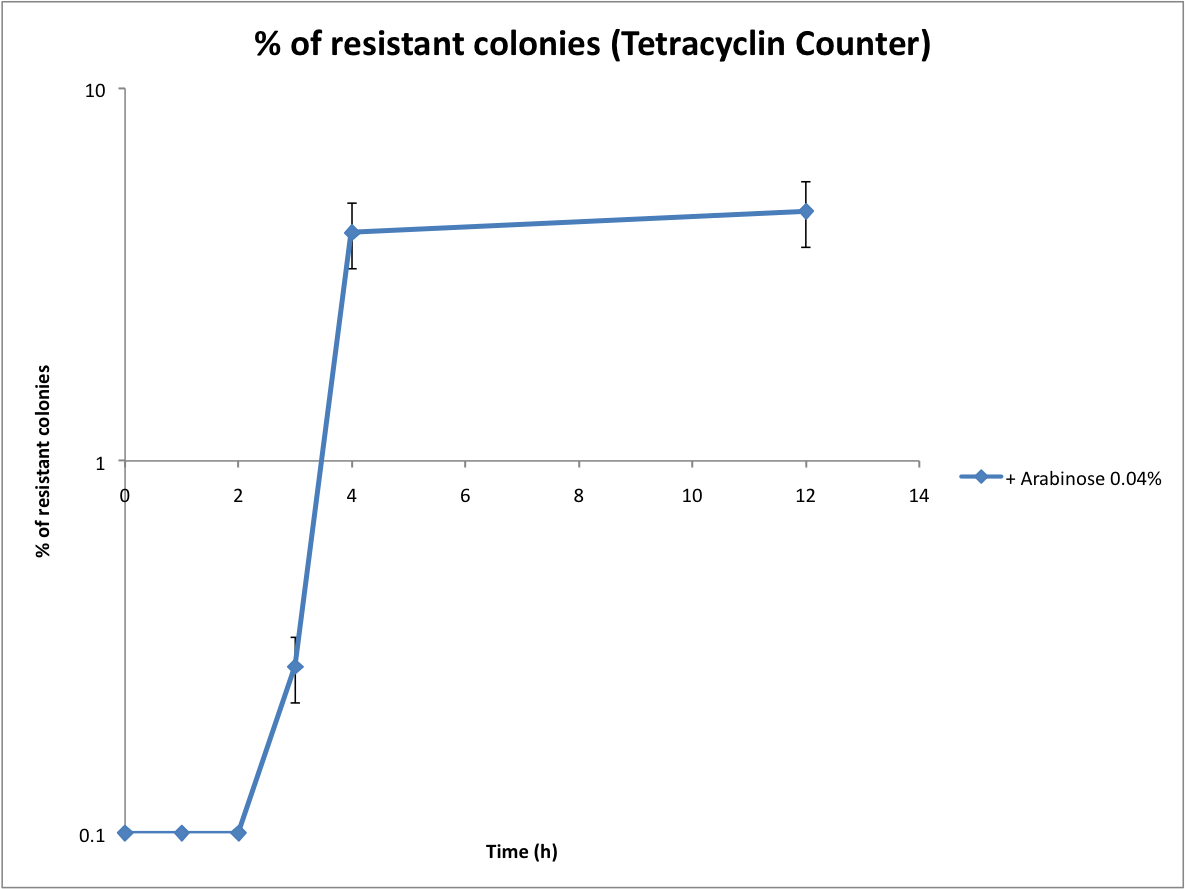
| - | == | + | ==Proof that the Tetracyline Counter works and first evaluation of the recombination rate== |
| - | + | ||
| - | + | <br>Following the same protocol, we tested the Tetracycline Counter. Only one copy of TetA(C) expressed is sufficient to induce tetracycline resistance. Therefore the number of resistant colonies is a good approximation of the recombination rate. | |
<br>[[Image:Tetracyclin resistance.jpg|centre|600px]] | <br>[[Image:Tetracyclin resistance.jpg|centre|600px]] | ||
| - | + | ||
<br><br> | <br><br> | ||
| - | + | ==Confirmation of the specific excision of the terminator by sequencing== | |
| + | <br> We took clones resistant to tetracycline from the past experiment, did overnight cultures and minipreps. Then we sent our minipreps for sequencing to confirm that IntI1 specifically cut the double terminator. Here you can see the nucleotide alignments. | ||
<html> | <html> | ||
<p style="display:block;"> | <p style="display:block;"> | ||
| Line 79: | Line 72: | ||
<img style="border:2px solid black" src="https://static.igem.org/mediawiki/2010/5/59/Unexcised_terminator-01.png" width="900px"> | <img style="border:2px solid black" src="https://static.igem.org/mediawiki/2010/5/59/Unexcised_terminator-01.png" width="900px"> | ||
</html> | </html> | ||
| + | |||
| + | |||
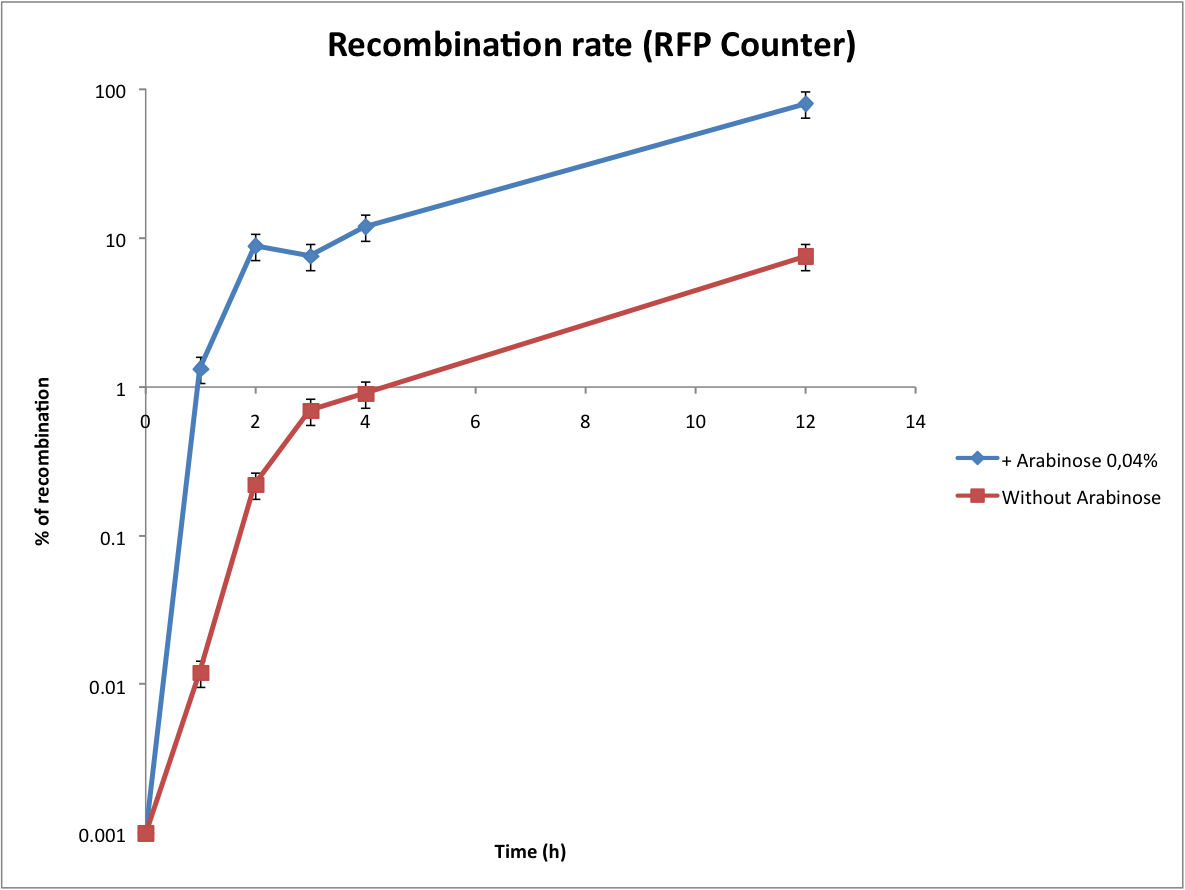
| + | ==Characterization of the recombination rate== | ||
| + | <html><p style="display:block;"> | ||
| + | At this point, it is important to determine the recombination rate of IntI1. To do this we have done our double transformations again on plates containing 1% glucose to inhibit the expression of IntI1. We did our overnight cultures with glucose for the same reason, then we diluted, waited until OD 0,2 and added arabinose 0,04%. As the counters are on high copy plasmids we found trick to calculate the recombination rate ! In every cell there are a lot of plasmids, some of them are recombined, others not. To have a good idea of this heterogeneity, we did minipreps at regular time intervals (in parallel, we directly plated the cultures). Then we transformed the minipreps that contain the whole population of plasmids. Statistically, only one plasmid can be integrated per competent cell. In this way, some cells are red (because they have only the excised plasmid) and other are not fluorescents (because they have only the non excised plasmid). This protocol is more complicated but it permits to significantly increase the sensitivity of the test (as you can see below). Unfortunately, the experiment with the Tetracycline Counter failed. The direct plating gives positive results but the transformation of the minipreps gives no colonies on tetracycline plates. After some researches we noted that the TetA(C) gene is toxic for the cells if over-expressed (see description of the biobrick). The principle of this experiment is to obtain only recombined plasmids in the cells, we can reasonably say that it would be lethal for the cells. | ||
| + | </html> | ||
| + | <br>[[Image:Recombination rate.jpg|centre|600px]] | ||
| + | |||
| + | From this graphs we can see the evolution of the recombination rate and logically the number of excised plasmids increases with time. If we want to count effectively, the recombination rate has to be really low (under 5%). From these results we can see that arabinose induction triggers too much recombinations. Actually the absence of arabinose is a perfect condition to test the counter (leaking of pBad). | ||
<br><br> | <br><br> | ||
| Line 85: | Line 87: | ||
<br><br> | <br><br> | ||
| - | ==Test the microfluidic device== | + | ==Test of the microfluidic device== |
bla | bla | ||
Revision as of 00:57, 28 October 2010
Construction of our counters
First of all, we spent a significant part of the summer to do cloning using the standard biobrick assembly protocol. For this, we have used a lot of biobricks submitted to the parts registry by previous teams, thank you ! Using these parts we have created plenty of new composite parts that are highly modular (different kind of RBS and promoters) and could serve next iGEM teams. We have also created some new biobricks that are at the core of our project. We hope other iGEM teams will work with integrons and might need these parts. We have constructed both the basic counter (with RFP and Tetracycline resistance gene) and the full counter + timer designs.
Moreover, each of our constructs was verified by sequencing. For more information you can take a look at the parts that we sent to the registry.
Proof that IntI1 integrase works !
Then we wanted to see if the IntI1 integrase actually works. We have done a double transformation "RFP Counter" (on pSB1A2 which is a high copy plasmid) + "pBAD-IntI1" (on pSulib which is a low copy plasmid with kanamycin resistance) to obtain a working counter. The protocol is quite simple : we dilute an overnight culture, wait until OD 0,2 and add arabinose to begin the test (t=0h). At regular time intervals, we plated on plates containing ampicillin (this is not important if the cells loose the "pBad-Int" plasmid because we don't want recombinations to happen on the plates).
This experiment shows us that IntI1 is effective ! The double terminator flanked by attC sites has been excised and the constitutive promoter upstream permits the expression of RFP. However, we have 2 important notes :
- At the beginning of the test, there already colonies that are fluorescents... We can explain this fact by the leaking of pBad that controls the expression of the integrase. Thereby the integrase is expressed at a low level and catalyzes the excision of the terminator. To overcome this problem we have to add glucose to the media which blocks the pBAD promoter and avoid recombination events to occur.
- The expression of RFP is inhomogeneous because the counter is on a high copy plasmid. So some cells have 2 recombined plasmids while others have 20 recombined plasmids ! To obtain a more linear response we have integrated our counter in a low copy plasmid (pSB4A5). In this way, there is more chance that only one event will happen in each cell.
Proof that the Tetracyline Counter works and first evaluation of the recombination rate
Following the same protocol, we tested the Tetracycline Counter. Only one copy of TetA(C) expressed is sufficient to induce tetracycline resistance. Therefore the number of resistant colonies is a good approximation of the recombination rate.
Confirmation of the specific excision of the terminator by sequencing
We took clones resistant to tetracycline from the past experiment, did overnight cultures and minipreps. Then we sent our minipreps for sequencing to confirm that IntI1 specifically cut the double terminator. Here you can see the nucleotide alignments.


Characterization of the recombination rate
At this point, it is important to determine the recombination rate of IntI1. To do this we have done our double transformations again on plates containing 1% glucose to inhibit the expression of IntI1. We did our overnight cultures with glucose for the same reason, then we diluted, waited until OD 0,2 and added arabinose 0,04%. As the counters are on high copy plasmids we found trick to calculate the recombination rate ! In every cell there are a lot of plasmids, some of them are recombined, others not. To have a good idea of this heterogeneity, we did minipreps at regular time intervals (in parallel, we directly plated the cultures). Then we transformed the minipreps that contain the whole population of plasmids. Statistically, only one plasmid can be integrated per competent cell. In this way, some cells are red (because they have only the excised plasmid) and other are not fluorescents (because they have only the non excised plasmid). This protocol is more complicated but it permits to significantly increase the sensitivity of the test (as you can see below). Unfortunately, the experiment with the Tetracycline Counter failed. The direct plating gives positive results but the transformation of the minipreps gives no colonies on tetracycline plates. After some researches we noted that the TetA(C) gene is toxic for the cells if over-expressed (see description of the biobrick). The principle of this experiment is to obtain only recombined plasmids in the cells, we can reasonably say that it would be lethal for the cells.
From this graphs we can see the evolution of the recombination rate and logically the number of excised plasmids increases with time. If we want to count effectively, the recombination rate has to be really low (under 5%). From these results we can see that arabinose induction triggers too much recombinations. Actually the absence of arabinose is a perfect condition to test the counter (leaking of pBad).
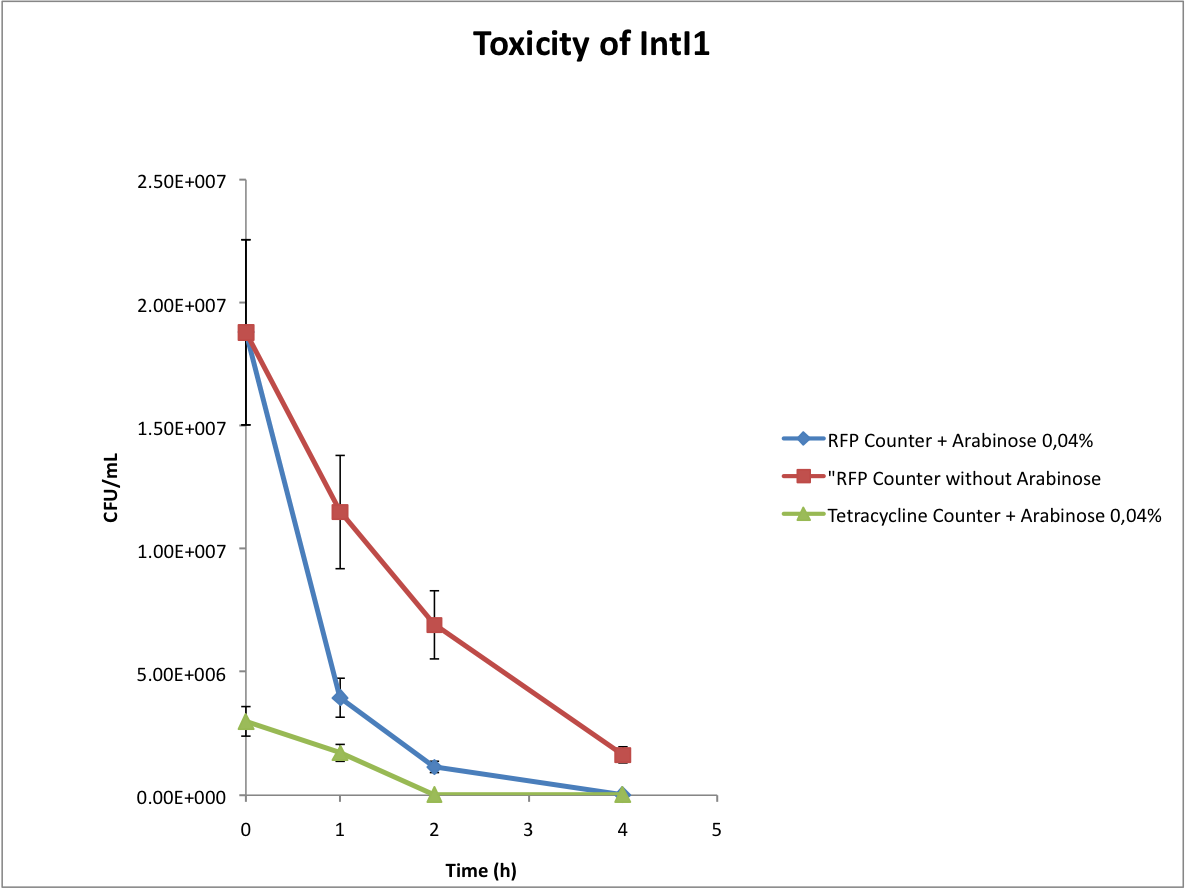
Verify the toxicity of the integrase
 "
"