Team:Osaka/Project cellulase
From 2010.igem.org
| (5 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
<p> | <p> | ||
| - | In order to create a practical system for Continuous Greening, our | + | In order to create a practical system for Continuous Greening, our microorganisms have to gain sustenance from dry, nutrient-poor soil. We turned to wood as an energy source, since can be decomposed by many kinds of microbes. Wood fibers are made of cellulose, hemicellulose, and lignin. Of those, cellulose comprises the largest proportion. Cellulose is made of β1→4 linked D-glucose units; therefore if we can break β-glycosidic bonds between the glucose units, our microbes can gain precious nutrients from dead wood in dry areas. Consequently, we focused on our efforts on realizing the degradation of cellulose through production and secretion of cellulase. Several different enzymes are involved in cellulose degradation and are collectively known as cellulases: endoglucanases break internal bonds in cellulose chains, exoglucanses break cellulose bonds near exposed ends of cellulose resulting in disaccharides called cellobiose, while β-glucocidase cleaves cellobiose molecules into 2 glucose units each. We aimed to produce all these enzymes from our microbe.</p> |
<p> | <p> | ||
| - | + | <h4>cellulose</h4> | |
| + | <img src="http://park11.wakwak.com/~nkon/misc/alcohol/cellulose3D.gif"> | ||
| + | </p> | ||
<p> | <p> | ||
| - | Today, our energy resources mostly depend on petroleum, coal, natural gas, and hydroelectricity. Some renewable energy such as sunlight, wind, rain, tides, and geothermal heal are known as renewable energy source; however, it only covers 19 % of global final energy consumption (2008). We need energy resource that does not depend on fossil fuel. If we can create cellulose degradation system, people can use the most abundant carbon resource in the world | + | In pursuance of more efficient degradation of cellulose, we investigated two different approaches. First, we BioBricked catalytic domains from many cellulases and tried combining them in various ways to obtain synergy between different parts. We theorized that combining the same set of genes in different orders will produce different proportions of each protein domain, thus resulting in differing efficiencies. We aimed to try several possible combinations and assay their activities. Second, we wanted looked into different microbes and their respective abilities to break down cellulose with the same set of cellulase parts. For this year’s project, we tried <i>Escherichia coli</i> and <i>Saccharomyces cerevisiae</i>.</p> |
| + | <p> | ||
| + | Today, our energy resources mostly depend on petroleum, coal, natural gas, and hydroelectricity. Some renewable energy such as sunlight, wind, rain, tides, and geothermal heal are known as renewable energy source; however, it only covers 19 % of global final energy consumption (2008). We need an energy resource that does not depend on fossil fuel. If we can create a cellulose degradation system, people can use the most abundant carbon resource in the world at a low cost. This will solve world problems such as energy and food consumption and contribute to energy sustainability.</p> | ||
<h3>BioBricks</h3> | <h3>BioBricks</h3> | ||
<p> | <p> | ||
| - | From BioBrick, we used endo-glucanase, cenA, (BBa_K118023)and exo-glucanase, cex, (BBa_K118022). Also, we used beta-gluctocidase which | + | From BioBrick, we used endo-glucanase, cenA, (BBa_K118023)and exo-glucanase, cex, (BBa_K118022). Also, we used beta-gluctocidase which the Edinburgh team kindly provided us. We initially cloned these genes into Silver fusion-compatible BioBricks. Furthermore, we cloned several other cellulase for new parts in BioBrick. To improve efficiency of cellulase, we also cloned cellulose binding module protein (BBa_K392014).In order to produce cellulase outside of microbe we used secretion tag, PelB(BBa_J32015)and OmpA(BBa_K103006). |
| - | + | ||
| - | + | ||
| - | + | ||
| + | <h3>Cellulose Binding Module</h3> | ||
| + | Cellulose and hemi-cellulose are insoluble; therefore, it is necessary to have close interaction between insoluble sugar chain and aqueous enzymes for optimal function. Cellulose binding modules (CBMs) are used in natural cellulolytic systems to enhance the activity of cellulases. We cloned several CBM in Silver fusion-compatible BioBricks for use in our cellulase constructs. | ||
| + | |||
| + | <h3>Secretion Tag</h3> | ||
| + | For secretion of cellulase outside of microbes, we inserted secretion tag in sequence. From Utah University, we got HlyA, TorA, and GeneIII for secretion tag of <i>Escherichia coli</i>. For <i>Saccharomyces cerevisiae</i>, Aglutinine 2 and sec tag were presented from New York University and we tried to clone alfa-aglutinine1 and suc tag by own. | ||
| + | |||
<h3>New Cellulase Parts</h3> | <h3>New Cellulase Parts</h3> | ||
| - | We cloned several new cellulase parts for BioBrick. These enzymes are Cel44A, Cel5B, Mananes, and xylanase. | + | We cloned several new cellulase parts for BioBrick. These enzymes are Cel44A, Cel5B, Mananes, and xylanase. |
<h3>Testing the cellulase</h3> | <h3>Testing the cellulase</h3> | ||
| Line 25: | Line 32: | ||
In nature, cellulose exists as insoluble compound. Therefore, in addition to pure cellulose, we used ASC and CMC as substrates to test for cellulase activity. ASC stands for Acid Swollen Cellulose and it pretreated with condensed phosphate. CMC is an abbreviation for carboxymethylcellulose and it is a derivative of cellulose. The structure of CMC is given below; a hydroxyl group is substituted for a carboxy-methyl group:</p> | In nature, cellulose exists as insoluble compound. Therefore, in addition to pure cellulose, we used ASC and CMC as substrates to test for cellulase activity. ASC stands for Acid Swollen Cellulose and it pretreated with condensed phosphate. CMC is an abbreviation for carboxymethylcellulose and it is a derivative of cellulose. The structure of CMC is given below; a hydroxyl group is substituted for a carboxy-methyl group:</p> | ||
<p> | <p> | ||
| - | <img src="http://upload.wikimedia.org/wikipedia/commons/d/d6/Carboxymethyl_cellulose.png"> | + | <h4>CMC</h4> |
| + | <img src="http://upload.wikimedia.org/wikipedia/commons/d/d6/Carboxymethyl_cellulose.png" width="150"> | ||
</p> | </p> | ||
Latest revision as of 03:38, 28 October 2010
Cellulase
In order to create a practical system for Continuous Greening, our microorganisms have to gain sustenance from dry, nutrient-poor soil. We turned to wood as an energy source, since can be decomposed by many kinds of microbes. Wood fibers are made of cellulose, hemicellulose, and lignin. Of those, cellulose comprises the largest proportion. Cellulose is made of β1→4 linked D-glucose units; therefore if we can break β-glycosidic bonds between the glucose units, our microbes can gain precious nutrients from dead wood in dry areas. Consequently, we focused on our efforts on realizing the degradation of cellulose through production and secretion of cellulase. Several different enzymes are involved in cellulose degradation and are collectively known as cellulases: endoglucanases break internal bonds in cellulose chains, exoglucanses break cellulose bonds near exposed ends of cellulose resulting in disaccharides called cellobiose, while β-glucocidase cleaves cellobiose molecules into 2 glucose units each. We aimed to produce all these enzymes from our microbe.
cellulose
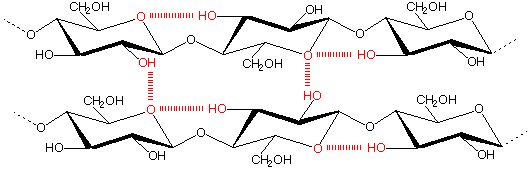
In pursuance of more efficient degradation of cellulose, we investigated two different approaches. First, we BioBricked catalytic domains from many cellulases and tried combining them in various ways to obtain synergy between different parts. We theorized that combining the same set of genes in different orders will produce different proportions of each protein domain, thus resulting in differing efficiencies. We aimed to try several possible combinations and assay their activities. Second, we wanted looked into different microbes and their respective abilities to break down cellulose with the same set of cellulase parts. For this year’s project, we tried Escherichia coli and Saccharomyces cerevisiae.
Today, our energy resources mostly depend on petroleum, coal, natural gas, and hydroelectricity. Some renewable energy such as sunlight, wind, rain, tides, and geothermal heal are known as renewable energy source; however, it only covers 19 % of global final energy consumption (2008). We need an energy resource that does not depend on fossil fuel. If we can create a cellulose degradation system, people can use the most abundant carbon resource in the world at a low cost. This will solve world problems such as energy and food consumption and contribute to energy sustainability.
BioBricks
From BioBrick, we used endo-glucanase, cenA, (BBa_K118023)and exo-glucanase, cex, (BBa_K118022). Also, we used beta-gluctocidase which the Edinburgh team kindly provided us. We initially cloned these genes into Silver fusion-compatible BioBricks. Furthermore, we cloned several other cellulase for new parts in BioBrick. To improve efficiency of cellulase, we also cloned cellulose binding module protein (BBa_K392014).In order to produce cellulase outside of microbe we used secretion tag, PelB(BBa_J32015)and OmpA(BBa_K103006).
Cellulose Binding Module
Cellulose and hemi-cellulose are insoluble; therefore, it is necessary to have close interaction between insoluble sugar chain and aqueous enzymes for optimal function. Cellulose binding modules (CBMs) are used in natural cellulolytic systems to enhance the activity of cellulases. We cloned several CBM in Silver fusion-compatible BioBricks for use in our cellulase constructs.Secretion Tag
For secretion of cellulase outside of microbes, we inserted secretion tag in sequence. From Utah University, we got HlyA, TorA, and GeneIII for secretion tag of Escherichia coli. For Saccharomyces cerevisiae, Aglutinine 2 and sec tag were presented from New York University and we tried to clone alfa-aglutinine1 and suc tag by own.New Cellulase Parts
We cloned several new cellulase parts for BioBrick. These enzymes are Cel44A, Cel5B, Mananes, and xylanase.Testing the cellulase
In nature, cellulose exists as insoluble compound. Therefore, in addition to pure cellulose, we used ASC and CMC as substrates to test for cellulase activity. ASC stands for Acid Swollen Cellulose and it pretreated with condensed phosphate. CMC is an abbreviation for carboxymethylcellulose and it is a derivative of cellulose. The structure of CMC is given below; a hydroxyl group is substituted for a carboxy-methyl group:
CMC

 "
"
