Team:Northwestern/Project/Modeling
From 2010.igem.org
(→Equations) |
|||
| Line 202: | Line 202: | ||
* dCC = kCf*E*Si - kCr*CC - kP*CC | * dCC = kCf*E*Si - kCr*CC - kP*CC | ||
* dP = kP*CC - kPdeg*P | * dP = kP*CC - kPdeg*P | ||
| + | |||
==='''IPTG Diffusion'''=== | ==='''IPTG Diffusion'''=== | ||
Revision as of 22:55, 25 October 2010
| Home | Brainstorm | Team | Acknowledgements | Project | Human Practices | Parts | Notebook | Calendar | Protocol | Safety | Links | References | Media | Contact |
|---|
|
| |||||||||||
ObjectiveThe main purpose of modeling was to characterize the topography of the kinetics behind foreign protein/product production in recombinant E.Coli biofilm with a diffusing inducer, and the effect on the various species involved by primarily the following factors:
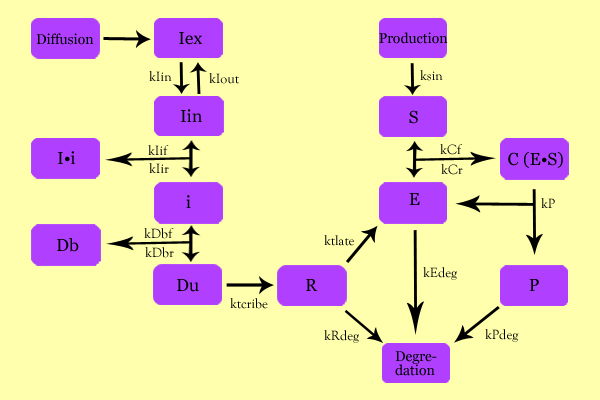
ModelingOverall ModelUsing enzyme kinetics equations, we elected to mathematically simulate the following model:
Variables
EquationsThe differential of the variables were found as follows:
IPTG DiffusionFirst, Fick's Law of Diffusion was modeled through MATLAB. The diffusion constant used was 220um^2/s.[4] IPTG was sprayed at the top of the colony, which then diffuses as according to Fick's law. The spatial local concentration will then differentially induce downstream processes.
Semi-Empirical DeterminationStatus: Under Development The initial plan was to use lacI-constitutive expression / lac-operon (CP-LacpI) part with Green Fluorescent Protein to acquire empirical data. By testing various combinations of CP/LacpI, RBS, and IPTG concentrations with GFP, one could acquire fluorescence data over time using a plate reader, and use that data to fit the current model, where instead of producing Chitin Synthase, the protein product would be GFP, the concentration of which could be measured as fluorescence. However, at the time of the wiki-freeze, data acquisition is incomplete.
Fitted ModelMatlab was used to generate a theoretical model where IPTG would diffuse down the biofilm as according to Fick's Law of Diffusion and initiate the process. The extracellular substrate concentration was assumed to be much greater than the uptake/use, and so would diffuse in at a constant rate. This model was fitted with empirical data using cp-lacpi-gfp to estimate the rate constant, and therefore the effects of varying cp, lacpi, and rbs on enzyme and final product production.
References1. A novel structured kinetic modeling approach for the analysis of plasmid instability in recombinant bacterial cultures William E. Bentley, Dhinakar S. Kompala Article first published online: 18 FEB 2004 DOI: 10.1002/bit.260330108 http://onlinelibrary.wiley.com/doi/10.1002/bit.260330108/pdf
Jongdae Lee, W. Fred Ramirez Article first published online: 19 FEB 2004 DOI: 10.1002/bit.260390608 http://onlinelibrary.wiley.com/doi/10.1002/bit.260390608/pdf
DOMINIQUE MENGIN-LECREULX, BERNARD FLOURET, AND JEAN VAN HEIJENOORT* E.R. 245 du C.N.R.S., Institut de Biochimie, Universit' Paris-Sud, Orsay, 91405, France Received 9 February 1983/Accepted 15 March 1983 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC217602/pdf/jbacter00247-0262.pdf
Philip S. Stewart Center for Biofilm Engineering and Department of Chemical Engineering, Montana State University–Bozeman, Bozeman, Montana, 59717-3980 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC148055/pdf/0965.pdf
PATRICIA L. EDELMANN' AND GORDON EDLIN Department of Genetics, University of California, Davis, California 95616 Received for publication 21 March 1974 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC245824/pdf/jbacter00335-0105.pdf
MATLAB file provided upon request. | |||||||||||
 "
"