Team:Northwestern/Project/Modeling
From 2010.igem.org
(→Modeling) |
|||
| Line 202: | Line 202: | ||
'''A novel structured kinetic modeling approach for the analysis of plasmid instability in recombinant bacterial cultures''' | '''A novel structured kinetic modeling approach for the analysis of plasmid instability in recombinant bacterial cultures''' | ||
| + | |||
William E. Bentley, Dhinakar S. Kompala | William E. Bentley, Dhinakar S. Kompala | ||
Article first published online: 18 FEB 2004 | Article first published online: 18 FEB 2004 | ||
DOI: 10.1002/bit.260330108 | DOI: 10.1002/bit.260330108 | ||
http://onlinelibrary.wiley.com/doi/10.1002/bit.260330108/pdf | http://onlinelibrary.wiley.com/doi/10.1002/bit.260330108/pdf | ||
| + | |||
'''Mathematical modeling of induced foreign protein production by recombinant bacteria''' | '''Mathematical modeling of induced foreign protein production by recombinant bacteria''' | ||
| + | |||
Jongdae Lee, W. Fred Ramirez | Jongdae Lee, W. Fred Ramirez | ||
Article first published online: 19 FEB 2004 | Article first published online: 19 FEB 2004 | ||
DOI: 10.1002/bit.260390608 | DOI: 10.1002/bit.260390608 | ||
http://onlinelibrary.wiley.com/doi/10.1002/bit.260390608/pdf | http://onlinelibrary.wiley.com/doi/10.1002/bit.260390608/pdf | ||
| + | |||
'''Pool Levels of UDP N-Acetylglucosamine and UDP NAcetylglucosamine-Enolpyruvate in Escherichia coli and Correlation with Peptidoglycan Synthesis''' | '''Pool Levels of UDP N-Acetylglucosamine and UDP NAcetylglucosamine-Enolpyruvate in Escherichia coli and Correlation with Peptidoglycan Synthesis''' | ||
| + | |||
DOMINIQUE MENGIN-LECREULX, BERNARD FLOURET, AND JEAN VAN HEIJENOORT* | DOMINIQUE MENGIN-LECREULX, BERNARD FLOURET, AND JEAN VAN HEIJENOORT* | ||
E.R. 245 du C.N.R.S., Institut de Biochimie, Universit' Paris-Sud, Orsay, 91405, France | E.R. 245 du C.N.R.S., Institut de Biochimie, Universit' Paris-Sud, Orsay, 91405, France | ||
Received 9 February 1983/Accepted 15 March 1983 | Received 9 February 1983/Accepted 15 March 1983 | ||
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC217602/pdf/jbacter00247-0262.pdf | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC217602/pdf/jbacter00247-0262.pdf | ||
| + | |||
'''Diffusion in Biofilms''' | '''Diffusion in Biofilms''' | ||
| + | |||
Philip S. Stewart | Philip S. Stewart | ||
Center for Biofilm Engineering and Department of Chemical Engineering, Montana | Center for Biofilm Engineering and Department of Chemical Engineering, Montana | ||
State University–Bozeman, Bozeman, Montana, 59717-3980 | State University–Bozeman, Bozeman, Montana, 59717-3980 | ||
http://www.ncbi.nlm.nih.gov/pmc/articles/PMC148055/pdf/0965.pdf | http://www.ncbi.nlm.nih.gov/pmc/articles/PMC148055/pdf/0965.pdf | ||
| + | |||
'''Regulation of the Synthesis of the Lactose Repressor''' | '''Regulation of the Synthesis of the Lactose Repressor''' | ||
| + | |||
PATRICIA L. EDELMANN' AND GORDON EDLIN | PATRICIA L. EDELMANN' AND GORDON EDLIN | ||
Department of Genetics, University of California, Davis, California 95616 | Department of Genetics, University of California, Davis, California 95616 | ||
Revision as of 23:25, 23 October 2010
| Home | Brainstorm | Team | Acknowledgements | Project | Human Practices | Parts | Notebook | Calendar | Protocol | Safety | Links | References | Media | Contact |
|---|
|
| |||||||||||
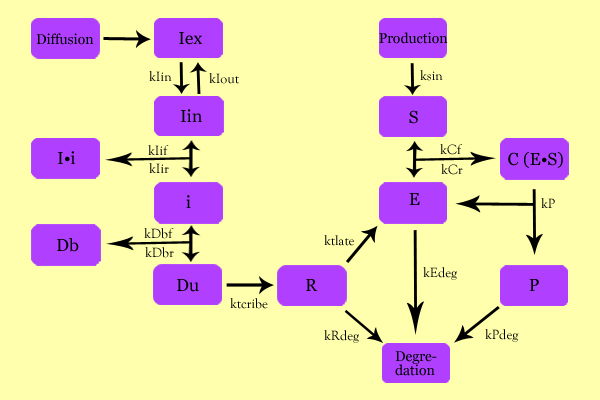
ModelingStatus: Under Development The purpose of Modeling was to characterize expression vectors in terms of expression level and time-delay, all activated by a diffusing inducer. Using simple enzyme kinetics equations, we elected to mathematically simulate the following model: Variables: Iex: External Inducer, determined by diffusion through Fick's law (IPTG in our experiment) Iin: Internal Inducer (IPTG) Ii: Inducer bound to Repressor (IPTG bound to lacI) i: Repressor (lacI) Db: Repressor-bound DNA (lacI-bound DNA(CHS3) region in plasmid) Dunb: transcribe-able or Repressor-unbound DNA (lacI-unbound DNA(CHS3)) Re: mRNA for Enzyme (CHS3 mRNA) E: Enzyme (CHS3) S: Substrate (N-Acetyl Glucosamine) C: Enzyme Substrate Complex (CHS3-(N-Acetyl-Glucosamine)-Chitin or (NAG)n Complex) P: Protein Product (Chitin or (NAG)n+1)
This model was fitted with empirical data using cp-lacpi-gfp to estimate the rate constant, and therefore the effects of varying cp, lacpi, and rbs on enzyme and final product production.
MATLAB file provided upon request.
A novel structured kinetic modeling approach for the analysis of plasmid instability in recombinant bacterial cultures William E. Bentley, Dhinakar S. Kompala Article first published online: 18 FEB 2004 DOI: 10.1002/bit.260330108 http://onlinelibrary.wiley.com/doi/10.1002/bit.260330108/pdf
Jongdae Lee, W. Fred Ramirez Article first published online: 19 FEB 2004 DOI: 10.1002/bit.260390608 http://onlinelibrary.wiley.com/doi/10.1002/bit.260390608/pdf
DOMINIQUE MENGIN-LECREULX, BERNARD FLOURET, AND JEAN VAN HEIJENOORT* E.R. 245 du C.N.R.S., Institut de Biochimie, Universit' Paris-Sud, Orsay, 91405, France Received 9 February 1983/Accepted 15 March 1983 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC217602/pdf/jbacter00247-0262.pdf
Philip S. Stewart Center for Biofilm Engineering and Department of Chemical Engineering, Montana State University–Bozeman, Bozeman, Montana, 59717-3980 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC148055/pdf/0965.pdf
PATRICIA L. EDELMANN' AND GORDON EDLIN Department of Genetics, University of California, Davis, California 95616 Received for publication 21 March 1974 http://www.ncbi.nlm.nih.gov/pmc/articles/PMC245824/pdf/jbacter00335-0105.pdf | |||||||||||
 "
"