Team:Northwestern/Project/Modeling
From 2010.igem.org
Timsterxox (Talk | contribs) |
(→Modeling) |
||
| Line 154: | Line 154: | ||
=='''Modeling'''== | =='''Modeling'''== | ||
| + | |||
| + | Status: Under Development | ||
| + | |||
| + | The purpose of Modeling was to characterize expression vectors in terms of expression level and time-delay, all activated by a diffused inducer. | ||
| + | |||
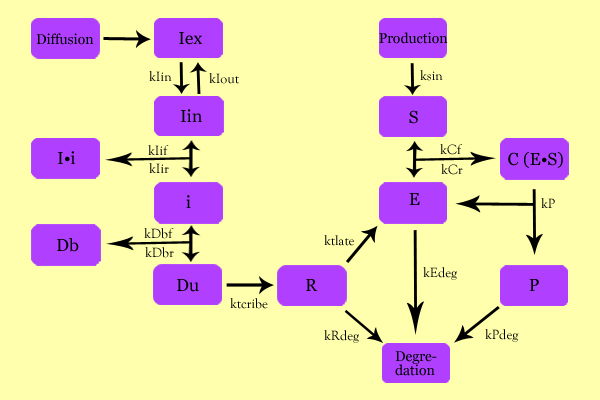
Using simple enzyme kinetics equations, we elected to mathematically simulate the following model: | Using simple enzyme kinetics equations, we elected to mathematically simulate the following model: | ||
[[Image:SuperModeling.jpg|500px|center]] | [[Image:SuperModeling.jpg|500px|center]] | ||
| - | |||
| - | Matlab was used to generate a theoretical model where IPTG would diffuse down the biofilm as according to Fick's Law of Diffusion. | + | Variables: |
| + | Iex: External Inducer, determined by diffusion through Fick's law (IPTG in our experiment) | ||
| + | Iin: Internal Inducer (IPTG) | ||
| + | Ii: Inducer bound to Repressor (IPTG bound to lacI) | ||
| + | i: Repressor (lacI) | ||
| + | Db: Repressor-bound DNA (lacI-bound DNA(CHS3) region in plasmid) | ||
| + | Dunb: transcribe-able or Repressor-unbound DNA (lacI-unbound DNA(CHS3)) | ||
| + | Re: mRNA for Enzyme (CHS3 mRNA) | ||
| + | E: Enzyme (CHS3) | ||
| + | S: Substrate (N-Acetyl Glucosamine) | ||
| + | C: Enzyme Substrate Complex (CHS3-(N-Acetyl-Glucosamine)-Chitin or (NAG)n Complex) | ||
| + | P: Protein Product (Chitin or (NAG)n+1) | ||
| + | |||
| + | Matlab was used to generate a theoretical model where IPTG would diffuse down the biofilm as according to Fick's Law of Diffusion and initiate the process. | ||
The extracellular substrate concentration was assumed to be much greater than the uptake/use, and so would diffuse in at a constant rate. | The extracellular substrate concentration was assumed to be much greater than the uptake/use, and so would diffuse in at a constant rate. | ||
This model was fitted with empirical data using cp-lacpi-gfp to estimate the effect of varying cp, lacpi, and rbs on enzyme and final product production. | This model was fitted with empirical data using cp-lacpi-gfp to estimate the effect of varying cp, lacpi, and rbs on enzyme and final product production. | ||
| + | |||
| + | References: | ||
| + | A novel structured kinetic modeling approach for the analysis of plasmid instability in recombinant bacterial cultures | ||
| + | William E. Bentley, Dhinakar S. Kompala | ||
| + | Article first published online: 18 FEB 2004 | ||
| + | |||
| + | DOI: 10.1002/bit.260330108 | ||
| + | http://onlinelibrary.wiley.com/doi/10.1002/bit.260330108/pdf | ||
| + | |||
| + | Mathematical modeling of induced foreign protein production by recombinant bacteria | ||
| + | Jongdae Lee, W. Fred Ramirez | ||
| + | Article first published online: 19 FEB 2004 | ||
| + | |||
| + | DOI: 10.1002/bit.260390608 | ||
| + | http://onlinelibrary.wiley.com/doi/10.1002/bit.260390608/pdf | ||
|} | |} | ||
Revision as of 23:15, 23 October 2010
| Home | Brainstorm | Team | Acknowledgements | Project | Human Practices | Parts | Notebook | Calendar | Protocol | Safety | Links | References | Media | Contact |
|---|
|
| |||||||||||
ModelingStatus: Under Development The purpose of Modeling was to characterize expression vectors in terms of expression level and time-delay, all activated by a diffused inducer. Using simple enzyme kinetics equations, we elected to mathematically simulate the following model: Variables: Iex: External Inducer, determined by diffusion through Fick's law (IPTG in our experiment) Iin: Internal Inducer (IPTG) Ii: Inducer bound to Repressor (IPTG bound to lacI) i: Repressor (lacI) Db: Repressor-bound DNA (lacI-bound DNA(CHS3) region in plasmid) Dunb: transcribe-able or Repressor-unbound DNA (lacI-unbound DNA(CHS3)) Re: mRNA for Enzyme (CHS3 mRNA) E: Enzyme (CHS3) S: Substrate (N-Acetyl Glucosamine) C: Enzyme Substrate Complex (CHS3-(N-Acetyl-Glucosamine)-Chitin or (NAG)n Complex) P: Protein Product (Chitin or (NAG)n+1) Matlab was used to generate a theoretical model where IPTG would diffuse down the biofilm as according to Fick's Law of Diffusion and initiate the process. The extracellular substrate concentration was assumed to be much greater than the uptake/use, and so would diffuse in at a constant rate. This model was fitted with empirical data using cp-lacpi-gfp to estimate the effect of varying cp, lacpi, and rbs on enzyme and final product production. References: A novel structured kinetic modeling approach for the analysis of plasmid instability in recombinant bacterial cultures William E. Bentley, Dhinakar S. Kompala Article first published online: 18 FEB 2004 DOI: 10.1002/bit.260330108 http://onlinelibrary.wiley.com/doi/10.1002/bit.260330108/pdf Mathematical modeling of induced foreign protein production by recombinant bacteria Jongdae Lee, W. Fred Ramirez Article first published online: 19 FEB 2004 DOI: 10.1002/bit.260390608 http://onlinelibrary.wiley.com/doi/10.1002/bit.260390608/pdf | |||||||||||
 "
"