Team:Newcastle/Urease
From 2010.igem.org
(→Calcium carbonate precipitation via urease expression) |
Swoodhouse (Talk | contribs) |
||
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| + | |||
| + | =Calcium carbonate precipitation via urease expression= | ||
| + | |||
| + | By increasing urease, precipitate calcium carbonate... | ||
| + | |||
| + | * The idea is to increase production of urease by | ||
| + | |||
| + | Biochemical network diagram.. | ||
| + | |||
| + | Computational model.. | ||
This idea of this biobrick is to eventually increase the production of calcium carbonate, by increasing the production of urease, indirectly. In order to do this, we have designed 2 biobricks; RocF and SR1. RocF is the gene that codes for the enzyme arginase, which in turn breaks down arginine to production urea, as well as ornithine. SR1, on the other hand acts as an antisense mRNA to AhrC mRNA | This idea of this biobrick is to eventually increase the production of calcium carbonate, by increasing the production of urease, indirectly. In order to do this, we have designed 2 biobricks; RocF and SR1. RocF is the gene that codes for the enzyme arginase, which in turn breaks down arginine to production urea, as well as ornithine. SR1, on the other hand acts as an antisense mRNA to AhrC mRNA | ||
| - | + | ||
| + | |||
UreA was efficiently expressed on the spore coat of ''B. subtilis'' when fused to CotB, CotC or CotG. Of these three coat proteins CotC allows the highest efficiency of expression, whereas CotB is the most appropriate for the display of heterologous proteins on the spore surface. | UreA was efficiently expressed on the spore coat of ''B. subtilis'' when fused to CotB, CotC or CotG. Of these three coat proteins CotC allows the highest efficiency of expression, whereas CotB is the most appropriate for the display of heterologous proteins on the spore surface. | ||
Revision as of 18:38, 25 June 2010

| |||||||||||||
| |||||||||||||
Calcium carbonate precipitation via urease expression
By increasing urease, precipitate calcium carbonate...
- The idea is to increase production of urease by
Biochemical network diagram..
Computational model..
This idea of this biobrick is to eventually increase the production of calcium carbonate, by increasing the production of urease, indirectly. In order to do this, we have designed 2 biobricks; RocF and SR1. RocF is the gene that codes for the enzyme arginase, which in turn breaks down arginine to production urea, as well as ornithine. SR1, on the other hand acts as an antisense mRNA to AhrC mRNA
UreA was efficiently expressed on the spore coat of B. subtilis when fused to CotB, CotC or CotG. Of these three coat proteins CotC allows the highest efficiency of expression, whereas CotB is the most appropriate for the display of heterologous proteins on the spore surface. Link: http://www.microbialcellfactories.com/content/pdf/1475-2859-9-2.pdf
Calcium Carbonate
Calcium carbonate precipitation is a widespread process among bacteria. The process are controlled by two distinct pathways, namely biologically induced pathway and the biologically controlled pathway. In the biologically controlled pathway, bacteria is able to control the precipitation of calcium carbonate and deposit it intracellularly. In contrast, in the biologically induced pathway, calcium carbonate are deposited extracellularly. No specialized cell structure or specific molecular mechanism is thought to be involved. However environmential conditions were known to play a part.
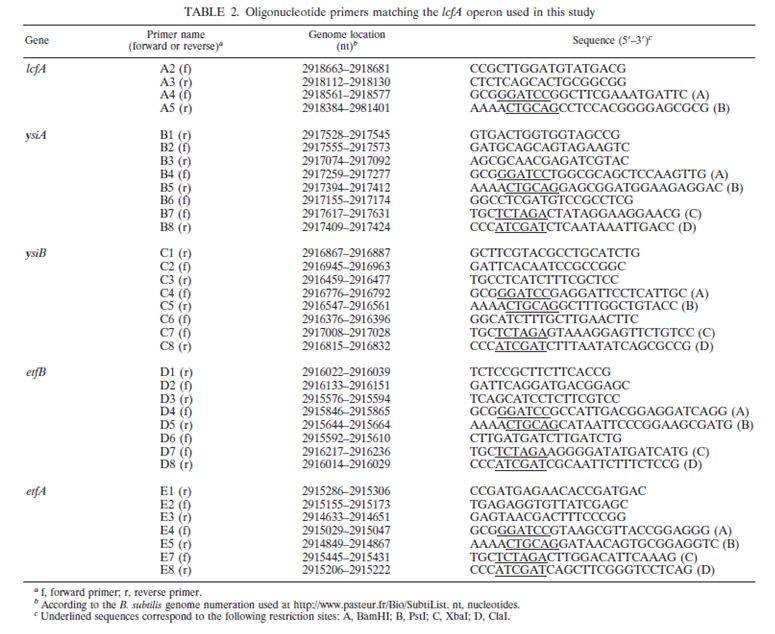
A cluster of five genes (lcfA, ysiA, ysiB, etfB, and etfA) called the lcfA operon that may involved in the calcium carbonate precipitation, as well as fatty acid metabolism in Bacillus subtilis. Analysis through inserional mutagenesis have shown that lcfA is not involved in calcium carbonate precipitation. Modulation of the lcfA operon using IPTG have pinpointed etfA to be essential, which encodes for a putative flavoprotein. Further analysis through RT-PCR have shown that lcfA to etfA gene is transcripted as a single transcription unit.
Reference: Barabesi, c., Galizzi, A., Mastromei, G., Rossi, M., Tamburini, E., and Perito, B. 2007. Bacillus subtilitis gene cluster involved in calcium carbonate biomineralization. j. Bacteriol. 189:228-235.
Recent report by Marvasi et al 2010 have shown that a decrease in the pH was the main process responsible for the lack of calcium carbonate precipitation (Barabesi et al 2007). This condition can be reversed when the biofilm was incubated in alkaline conditions. Therefore it has been hypothesize that a possible link exists between proton extrusion and the cytosolic accumulation of NADH in B.subtilis.
Reference: Marvasi, M., T.visscher, P., Perito, B., Mastromei, G., and Casillas-Mart´ınez, L. Physiological requirements for carbonate precipitation during biofilm development of Bacillus subtilis etfA mutant. FEMS. 71:341-350.
Biosynthesis of Active Bacillus subtilis Urease in the Absence of Known Urease Accessory Proteins JOURNAL OF BACTERIOLOGY, Oct. 2005, p. 7150–7154 Jong Kyong Kim, Scott B. Mulrooney, and Robert P. Hausinger Cement and Concrete Research 40 (2010) 157–166
Use of bacteria to repair cracks in concrete Kim Van Tittelboom a, Nele De Belie a,⁎, Willem De Muynck a,b, Willy Verstraete b
Expression of the rocDEF Operon Involved in Arginine Catabolism in Bacillus subtilis
Rozenn Gardan, Georges Rapoport and Michel Debarbouille J. Mol. Biol. (1995) 249, 843–856
Expression of the Bacillus subtilis ureABC Operon Is Controlled
by Multiple Regulatory Factors Including CodY,
GlnR, TnrA, and Spo0H
 
|
 "
"