Team:Newcastle/6 August 2010
From 2010.igem.org
(→Discussion) |
|||
| (49 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:Newcastle/mainbanner}} | {{Team:Newcastle/mainbanner}} | ||
| - | =Gel Electrophoresis for the | + | =Gel Electrophoresis for the amplified fragments of Pspac_oid promoter and ''lacI''= |
==Aim== | ==Aim== | ||
| - | The aim of the experiment is to perform gel electrophoresis for the two PCR reactions | + | The aim of the experiment is to perform gel electrophoresis for the two PCR reactions, ''lacI'' and Pspac_oid promoter that were done using a different stock of plasmid pMutin4 on 5th August, 2010. |
==Materials and Protocol== | ==Materials and Protocol== | ||
| Line 10: | Line 10: | ||
==Result== | ==Result== | ||
| + | |||
| + | [[Image:Newcastle_060810_gel_1.png|400px]] | ||
| + | |||
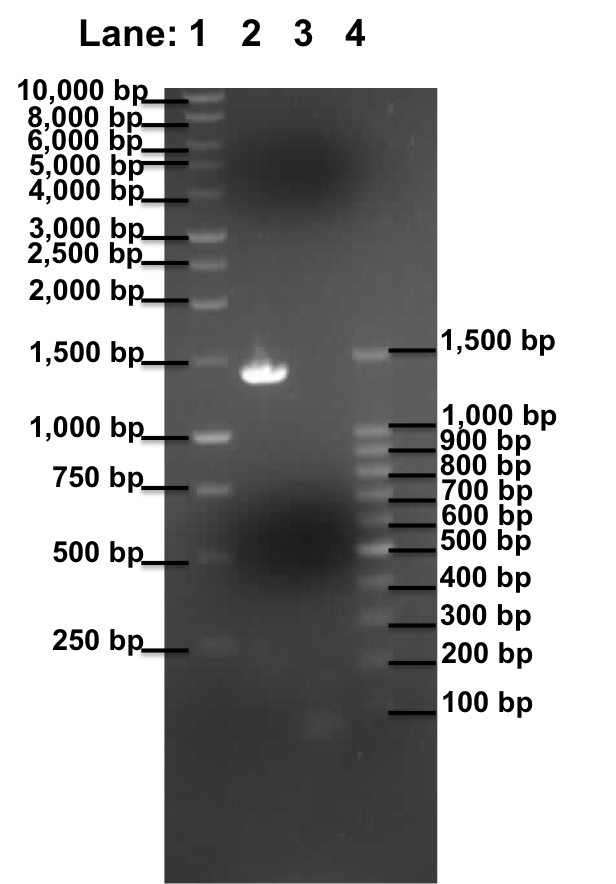
| + | '''Figure 1''': Gel electrophoresis of the ''lacI'' and Pspac_oid promoter. | ||
| + | |||
* '''Lane 1''': 1kb DNA ladder | * '''Lane 1''': 1kb DNA ladder | ||
* '''Lane 2''': Plamid pMutin4 containing ''lacI'' | * '''Lane 2''': Plamid pMutin4 containing ''lacI'' | ||
| Line 18: | Line 23: | ||
|- | |- | ||
! | ! | ||
| - | + | !'''Pspac_oid promoter''' | |
| - | !'''Pspac_oid | + | !'''''lacI''''' |
| - | !''' | + | |
| - | + | ||
| - | + | ||
| - | + | ||
|- | |- | ||
|'''Size of the Fragment (in bp)''' | |'''Size of the Fragment (in bp)''' | ||
| - | |||
|106 approx. | |106 approx. | ||
| - | | | + | |1400 approx. |
| - | + | ||
| - | + | ||
| - | + | ||
|} | |} | ||
'''Table 1''': Table represents the size of the fragments represented as bands on the gel in their corresponding lanes. | '''Table 1''': Table represents the size of the fragments represented as bands on the gel in their corresponding lanes. | ||
==Discussion== | ==Discussion== | ||
| - | + | A correct sized band was observed in lane 2, ''lacI'' which serve as the positive control. However no band was observed in lane 3, which contain the Pspac_oid promoter. | |
==Conclusion== | ==Conclusion== | ||
| - | This experiment shows that the | + | This experiment shows that the plasmid pMutin4 is intact as the ''lacI'' frangment have been successfully amplified (Lane 2). However the Pspac_oid promoter are still not being amplified. This could be due to the following reason: |
| - | + | ||
# Melting temperature could be incorrect. | # Melting temperature could be incorrect. | ||
| - | |||
| + | ==Solution for the problem== | ||
| + | # Try a range of melting temperature. | ||
| - | = | + | =Amplification of Pspac_oid promoter by PCR= |
| - | ==Aim== | + | ==Aim== |
| + | The aim of this experiment is to amplify the Pspac_oid promoter from plasmid pMutin4 for the construction of [[Team:Newcastle/Urease|''rocF'' BioBrick]] using Phusion PCR. | ||
| - | + | ==Materials and Protocol== | |
| + | Please refer to [[Team:Newcastle/PCR| PCR]] for Phusion PCR protocol. The details for the 2 PCR reactions are mentioned below: | ||
| - | == | + | ===PCR=== |
| + | {|border=1 | ||
| + | |- | ||
| + | !'''Tube''' | ||
| + | !'''Part to be amplified''' | ||
| + | !'''DNA fragment consisting the part''' | ||
| + | !'''Forward primer''' | ||
| + | !'''Reverse Primer''' | ||
| + | !'''Melting Temperature (Tm in °C) ''' | ||
| + | !'''Size of the fragment (in bp)''' | ||
| + | !'''Extension time* (in seconds)''' | ||
| + | |- | ||
| + | |1 | ||
| + | |Pspacoid Promoter | ||
| + | |pMutin4 | ||
| + | |P1P1 forward | ||
| + | |P2P1 reverse | ||
| + | |49 | ||
| + | |106 approx. | ||
| + | |15 | ||
| + | |- | ||
| + | |2 | ||
| + | |Pspacoid Promoter | ||
| + | |pMutin4 | ||
| + | |P1P1 forward | ||
| + | |P2P1 reverse | ||
| + | |50 | ||
| + | |106 approx. | ||
| + | |15 | ||
| + | |} | ||
| - | + | '''Table 2''': Table represents 2 different Phusion PCR reactions for the amplification of Pspac_oid promoter, so that it can be ligated together with other fragments for the construction of ''rocF'' with the help of Gibson Cloning method. | |
| + | * The extension rate of the Phusion polymerase is 1Kb/ 30 seconds. Therefore the extension time of each PCR reaction is different. | ||
| + | * To learn more about the ''rocF'' fragments, please refer to the [[Media:Cloning_strategy_for_rocF.pdf|Cloning strategy for ''rocF'']]. | ||
| - | == | + | ==Result== |
| + | [[Image:Newcastle_060810_gel_2.png|400px]] | ||
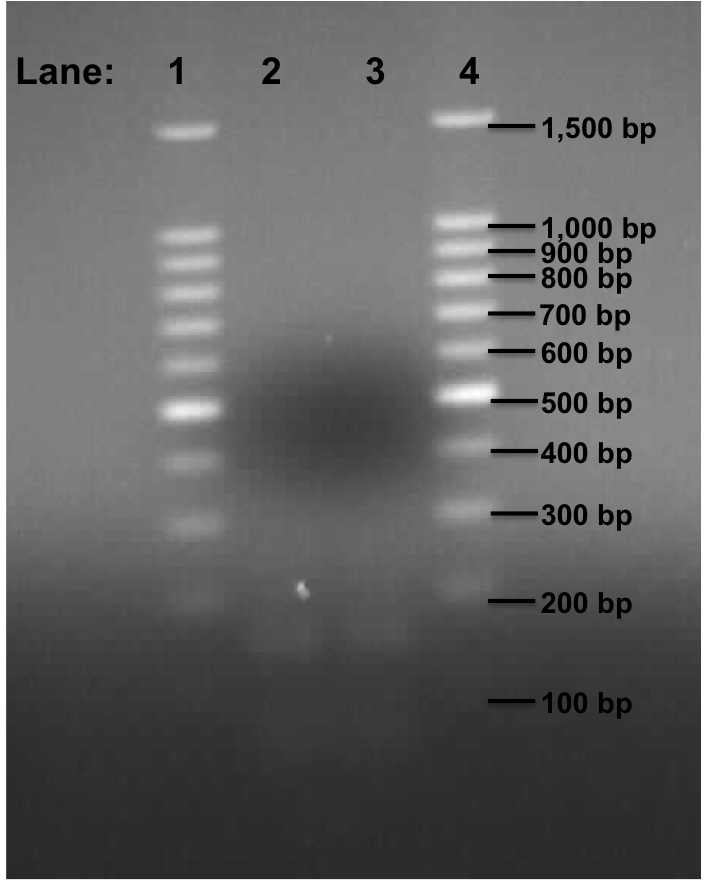
| - | + | '''Figure 2''': Gel electrophoresis of the Pspac_oid promoter from plasmid pMutin4. | |
| - | + | * '''Lane 1''': 100bp DNA ladder | |
| + | * '''Lane 2''': Plamid pMutin4 containing Pspac_oid promoter at 49°C | ||
| + | * '''Lane 3''': Plamid pMutin4 containing Pspac_oid promoter at 50°C | ||
| + | * '''Lane 4''': 100bp DNA ladder | ||
| - | + | {|border=1 | |
| - | + | |- | |
| - | + | ! | |
| - | + | !'''Pspac_oid promoter at 49°C''' | |
| - | + | !'''Pspac_oid promoter at 50°C''' | |
| - | + | |- | |
| - | + | |'''Size of the Fragment (in bp)''' | |
| - | + | |106 approx. | |
| - | + | |106 approx. | |
| - | + | |} | |
| - | + | '''Table 3''': Table represents the size of the fragments represented as bands on the gel in their corresponding lanes. | |
| - | + | ||
| - | + | ||
==Discussion== | ==Discussion== | ||
| - | + | We found two very faint bands in the lanes 2 and 3. | |
| - | We | + | |
==Conclusion== | ==Conclusion== | ||
| + | This experiment shows that the PCR reaction was unsuccessful even though we got a faint band for Pspac_oid promoter. This time, we are going to use plasmid containing KinA Biobrick and plasmid containing stochastic switch Biobrick which were devised by Team Newcastle 2009. We would be dealing with this on Monday 9th August, 2010. | ||
{{Team:Newcastle/footer}} | {{Team:Newcastle/footer}} | ||
Latest revision as of 23:24, 25 October 2010

| |||||||||||||
| |||||||||||||
Contents |
Gel Electrophoresis for the amplified fragments of Pspac_oid promoter and lacI
Aim
The aim of the experiment is to perform gel electrophoresis for the two PCR reactions, lacI and Pspac_oid promoter that were done using a different stock of plasmid pMutin4 on 5th August, 2010.
Materials and Protocol
Please refer to: Gel electrophoresis.
Result
Figure 1: Gel electrophoresis of the lacI and Pspac_oid promoter.
- Lane 1: 1kb DNA ladder
- Lane 2: Plamid pMutin4 containing lacI
- Lane 3: Plamid pMutin4 containing Pspac_oid promoter
- Lane 4: 100bp DNA ladder
| Pspac_oid promoter | lacI | |
|---|---|---|
| Size of the Fragment (in bp) | 106 approx. | 1400 approx. |
Table 1: Table represents the size of the fragments represented as bands on the gel in their corresponding lanes.
Discussion
A correct sized band was observed in lane 2, lacI which serve as the positive control. However no band was observed in lane 3, which contain the Pspac_oid promoter.
Conclusion
This experiment shows that the plasmid pMutin4 is intact as the lacI frangment have been successfully amplified (Lane 2). However the Pspac_oid promoter are still not being amplified. This could be due to the following reason:
- Melting temperature could be incorrect.
Solution for the problem
- Try a range of melting temperature.
Amplification of Pspac_oid promoter by PCR
Aim
The aim of this experiment is to amplify the Pspac_oid promoter from plasmid pMutin4 for the construction of rocF BioBrick using Phusion PCR.
Materials and Protocol
Please refer to PCR for Phusion PCR protocol. The details for the 2 PCR reactions are mentioned below:
PCR
| Tube | Part to be amplified | DNA fragment consisting the part | Forward primer | Reverse Primer | Melting Temperature (Tm in °C) | Size of the fragment (in bp) | Extension time* (in seconds) |
|---|---|---|---|---|---|---|---|
| 1 | Pspacoid Promoter | pMutin4 | P1P1 forward | P2P1 reverse | 49 | 106 approx. | 15 |
| 2 | Pspacoid Promoter | pMutin4 | P1P1 forward | P2P1 reverse | 50 | 106 approx. | 15 |
Table 2: Table represents 2 different Phusion PCR reactions for the amplification of Pspac_oid promoter, so that it can be ligated together with other fragments for the construction of rocF with the help of Gibson Cloning method.
- The extension rate of the Phusion polymerase is 1Kb/ 30 seconds. Therefore the extension time of each PCR reaction is different.
- To learn more about the rocF fragments, please refer to the Cloning strategy for rocF.
Result
Figure 2: Gel electrophoresis of the Pspac_oid promoter from plasmid pMutin4.
- Lane 1: 100bp DNA ladder
- Lane 2: Plamid pMutin4 containing Pspac_oid promoter at 49°C
- Lane 3: Plamid pMutin4 containing Pspac_oid promoter at 50°C
- Lane 4: 100bp DNA ladder
| Pspac_oid promoter at 49°C | Pspac_oid promoter at 50°C | |
|---|---|---|
| Size of the Fragment (in bp) | 106 approx. | 106 approx. |
Table 3: Table represents the size of the fragments represented as bands on the gel in their corresponding lanes.
Discussion
We found two very faint bands in the lanes 2 and 3.
Conclusion
This experiment shows that the PCR reaction was unsuccessful even though we got a faint band for Pspac_oid promoter. This time, we are going to use plasmid containing KinA Biobrick and plasmid containing stochastic switch Biobrick which were devised by Team Newcastle 2009. We would be dealing with this on Monday 9th August, 2010.
 
|
 "
"